- EN - English
- CN - 中文
Microdissection and Single-Cell Suspension of Neocortical Layers From Ferret Brain for Single-Cell Assays
灵长类大脑皮层层次微切割与单细胞悬浮液制备方法
发布: 2024年12月20日第14卷第24期 DOI: 10.21769/BioProtoc.5133 浏览次数: 2690
评审: Pilar Villacampa AlcubierreAnonymous reviewer(s)
Abstract
Brain development is highly complex and dynamic. During this process, the different brain structures acquire new components, such as the cerebral cortex, which builds up different germinal and cortical layers during its development. The genetic study of this complex structure has been commonly approached by bulk-sequencing of the entire cortex as a whole. Here, we describe the methodology to study this layered tissue in all its complexity by microdissecting two germinal layers at two developmental time points. This protocol is combined with a step-by-step explanation of tissue dissociation that provides high-quality cells ready to be analyzed by the newly developed single-cell assays, such as scRNA-seq, scATAC-seq, and TrackerSeq. Altogether, this approach increases the resolution of the genetic analyses from the cerebral cortex compared to bulk studies. It also facilitates the study of laboratory animal models that recapitulate human cortical development better than mice, like ferrets.
Key features
• Microdissection of individual germinal layers in the developing cerebral cortex from living brain slices.
• Enzymatic and mechanical dissociation generates single-cell suspensions available for high-throughput single-cell assays.
• Protocol optimized for embryonic and early postnatal ferret cortex.
Keywords: Cerebral cortex (大脑皮层)Graphical overview
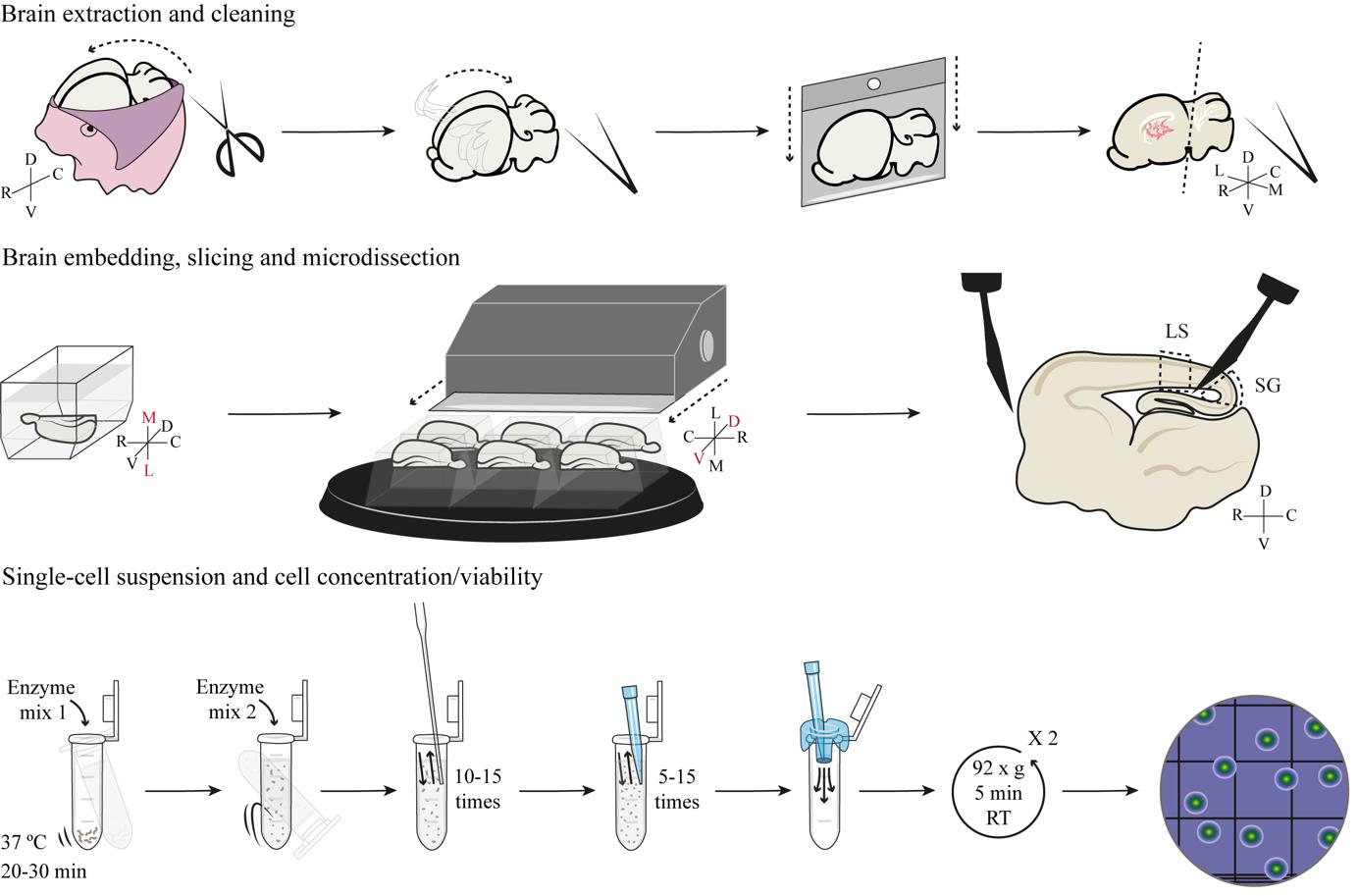
Background
The field of cerebral cortex development has witnessed milestone breakthroughs over the last 15 years, including the discovery of new types of progenitor cells such as basal radial glia (bRG) [1–5] and truncated radial glia (tRG) [6–8]. The discovery of new neural progenitor cell types has derived from more detailed characterizations of these cells, revealing for example that they dynamically modify their morphology along development, defining cell morphotypes [9–11]. The lineage relationships between this wide repertoire of progenitor cell morphotypes are also variable and dynamic, in line with the complexity of intrinsic and extrinsic factors regulating this process [9]. This complexity is greater in mammals with a folded cortex, like humans or ferrets, compared to species with a smooth cortex, like mice [8].
Traditionally, transcriptomic analyses of the developing cerebral cortex were possible only in bulk, considering it a relatively simple and homogeneous structure in spite of being clearly layered and anatomically subdivided. However, the emergence of single-cell sequencing technologies (i.e., scRNA-seq) has allowed the transcriptomic characterization of individual cell types with unprecedented detail and the discovery of unsuspected cell diversity also in the cerebral cortex. Nevertheless, scRNA-seq analyses of the developing cortex are still largely analyzed in bulk, relying on a limited number of specific, presumed marker genes to identify cell types. An increasing number of studies question if such marker genes are reliable for studying cell diversity in the developing cortex, particularly neural progenitor cells, and if they are faithful across mammalian species [8,9,12–15]. This protocol represents a milestone for studies of cortex development, where microdissection of individual layers of the developing cortex allows grasping its full complexity. By taking advantage of the extensive knowledge of the biology of cortical progenitor cells, the microdissection of germinal layers presented here allows distinguishing progenitor cell populations based on their layer of residency (i.e., apical versus basal progenitors), independent from the use of marker genes that must be assumed specific to particular cell types. Furthermore, the generation of a single-cell suspension from these microdissected tissues enables the use of massively parallel single-cell assays, including scRNA-seq or scATAC-seq, as well as TrackerSeq for cell lineage tracing. Hence, the combination of microdissection of individual germinal zones with single-cell approaches vastly increases the power of these analyses compared to studies of bulk tissue. Last, this protocol facilitates the study of animal models that are less common and more complex than mice, such as ferrets, with features of cortical development similar to humans. Unfortunately, the dissociation of cortical tissue into individual cells bears inherent limitations, such as the impossibility of cell morphology assessment, which currently limits the association of the new data with previous characterizations of progenitor cell morphotypes.
The specific details and conditions presented in this protocol have been optimized for the cerebral cortex of developing ferrets; nevertheless, it can also be implemented to study other layered brain regions, such as the hippocampus, olfactory bulb, or superior colliculus. The study of these structures by microdissection of individual layers, combined with the ever-increasing possibilities of single-cell assays, will significantly contribute to redefining cell type diversity in the brain.
Materials and reagents
Biological materials
Pregnant sable ferret (Mustela putorius furo) (Euroferrets, Denmark)
Reagents
Blue ultrasound gel (Transonic, catalog number: TOG04)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888)
Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S3264)
Potassium chloride (KCl) (VWR, catalog number: 26759.291)
Potassium dihydrogen phosphate (KH2PO4) (VWR, catalog number: 26925.295)
MACS neural tissue dissociation kit (P) (Miltenyi Biotec, catalog number: 130-092-628)
Leibovitz’s 1× L-15 medium with L-glutamine, without phenol red (Gibco, catalog number: 21083027)
SeaPlaque agarose (Lonza, catalog number: 50100)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906)
1× Phosphate buffered saline (PBS) without CaCl2/MgCl2 pH 7.4 (Gibco, catalog number: 10010023)
RNaseZap RNase decontamination solution (Invitrogen, catalog number: AM9780)
Anesthetic/analgesic cocktail: xylazine (Karizoo, trade name: Xylasol 20 mg/mL, catalog number: 579700.7), ketamine (VetViva Richter, trade name: Ketamidor 100 mg/mL, catalog number: 580393.7), buprenorphine (VetViva Richter, trade name: Bupaq 0.3 mg/mL, catalog number: 578816.6)
Saline solution (NaCl 0.9%) (Interapothek, catalog number: 208978)
1× Hank’s balanced salt solution (HBSS) without CaCl2/MgCl2/phenol red (Gibco, catalog number: 14175095)
Ethanol absolute (VWR, catalog number: 20821.365P)
0.4% Trypan blue stain (Invitrogen, catalog number: T10282)
Solutions
10× Phosphate buffered saline (PBS) stock solution (see Recipes)
1× PBS (see Recipes)
Cell collection medium (see Recipes)
Recipes
10× PBS stock solution (1 L)
Reagent Final concentration Quantity or Volume NaCl 1.37 M 80 g Na2HPO4 100 mM 14.4 g KCl 27 mM 2 g KH2PO4 18 mM 2.4 g Distilled H2O n/a 1 L* Total n/a 1 L *Dilute reagents in 800 mL, bring solution pH to 7.4, and top off to 1 L.
1× PBS (1 L)
Reagent Final concentration Quantity or Volume PBS 10× 1× 100 mL Distilled H2O n/a 900 mL Total n/a 1 L Cell collection medium (50 mL)
Reagent Final concentration Quantity or Volume BSA 0.04% (w/v) 0.02 g PBS (without CaCl2/MgCl2, pH 7.4) 1× 50 mL Total n/a 50 mL The medium should be freshly prepared and filtered on the day of the experiment.
Laboratory supplies
Glass Pasteur pipettes (Fisher Scientific, catalog number: FB50253)
0.5 mL PCR tubes (Eppendorf, catalog number: 0030124537)
2 mL DNA LoBind tubes (Eppendorf, catalog number: 0030108078)
15 mL centrifuge tubes (Corning, catalog number: 430791)
50 mL centrifuge tubes (Corning, catalog number: 430829)
40 μm PluriStrainer mini cell strainers (PluriSelect, catalog number: 43-10040-40)
0.22 μm syringe filter (VWR, catalog number: 514-1263)
50 mL sterile syringe (BD, catalog number: 300865)
Pipetman P1000 tips (Daslab, catalog number: 162222)
Pipetman P200 tips (Daslab, catalog number: 162001)
Pipetman P20 tips (Daslab, catalog number: 162001)
Permanent marker pen (Sarstedt, catalog number: 95.954)
100 mm × 15 mm sterilized Petri dishes (JetBiofil, catalog number: TCD010100)
Ice buckets
500 mL PYREX griffin low-form beaker (Corning, catalog number: 1000J-500)
1 mL sterile syringe (ENFA, catalog number: JS1)
10 mL sterile syringe (BD, catalog number: 307736)
Microlance 3 hypodermic needles 25 G × 5/8'', 16 × 0.5 mm (BD, catalog number: 300600)
Microlance 3 hypodermic needles 30 G × 1/2'', 13 × 0.3 mm (BD, catalog number: 304000)
Polypropylene laboratory tray (Vitlab, catalog number: 72298)
Single-edge razor blade (KBC, catalog number: 1-909-11036)
Double-edge razor blade (Gillette Platinum, catalog number: 21377003)
Peel-A-Way embedding molds truncated - T12 (Polysciences, catalog number: 18986-1)
Peel-A-Way embedding molds rectangular - R30 (Polysciences, catalog number: 18646B-1)
Super Glue-3 (Loctite, catalog number: 2640076)
LDPE Pasteur pipette (Fisher Scientific, catalog number: 747760)
20 μL ZEROTIP pipette micro tip with filter (JetBiofil, catalog number: PMT252020)
2 mL floating tube rack (Sigma-Aldrich, catalog number: R7776-5EA)
Digital timer (Scharlau, catalog number: 038-000566)
Pipetman P10 tips (Daslab, catalog number: 163030)
0.2 mL PCR tubes (Eppendorf, catalog number: 0030124332)
Hand tally counter (VWR, catalog number: 710-0935)
Countess cell counting chamber slides (Invitrogen, catalog number: C10228)
Equipment
Ultrasound system (SonoSite, model: MicroMaxx)
Distilled water (H2O) (Milli-Q, model: IQ-7000)
pH meter (Crison, model: Basic 20+)
Autoclave (Tuttnauer, model: 2540M)
Bunsen burner (Carl Roth, catalog number: CK29.1)
Drying oven (POL-EKO, model: SLW400)
Straight, sharp tips, fine dissecting scissors (Fine Science Tools, catalog number: 14060-09)
Straight, super fine tips dissecting forceps (Fine Science Tools, catalog number: Dumont #5SF 11252-00)
Micro spoon spatulas (RSG Solingen, catalog number: 80.025.150)
Micro flat-ended spatula (RSG Solingen, catalog number: 80.038.150)
15° straight, sharp pointed tips, microsurgical stab knives (MSP, catalog number: 72-1501)
Straight, sharp/blunt tips dissecting scissors (Fine Science Tools, catalog number: 14001-18)
Pipetman P1000 (Gilson, catalog number: F144059M)
Pipetman P200 (Gilson, catalog number: F144058M)
Pipetman P20 (Gilson, catalog number: F144056M)
Laminar flow hood (Telstar, model: CytoStar)
Water bath at 42 °C (MRC, catalog number: WBO-100)
Dissecting scope with incident illumination (Leica, model: M60)
Vibratome (Leica, model: VT1000 S)
Dissecting scope with transmitted illumination (Leica, model: MS5)
Vortex mixer (Labnet)
Water bath at 37 °C (MRC, catalog number: WBO-200)
Microcentrifuge (Labnet, model: Spectrafuge 24D)
Blaubrand Neubauer counting chamber (Brand, catalog number: 718605)
Pipetman P10 (Gilson, catalog number: F144055M)
Inverted microscope (Leica, model: DM IL)
Countess II FL automated cell counter (Invitrogen, catalog number: AMQAF1000)
Procedure
文章信息
稿件历史记录
提交日期: Jul 15, 2024
接收日期: Oct 5, 2024
在线发布日期: Oct 29, 2024
出版日期: Dec 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Del-Valle-Anton, L., Amin, S. and Borrell, V. (2024). Microdissection and Single-Cell Suspension of Neocortical Layers From Ferret Brain for Single-Cell Assays. Bio-protocol 14(24): e5133. DOI: 10.21769/BioProtoc.5133.
- Del-Valle-Anton, L., Amin, S., Cimino, D., Neuhaus, F., Dvoretskova, E., Fernández, V., Babal, Y. K., Garcia-Frigola, C., Prieto-Colomina, A., Murcia-Ramón, R., et al. (2024). Multiple parallel cell lineages in the developing mammalian cerebral cortex. Sci Adv. 10(13): eadn9998.
分类
神经科学 > 发育 > 形态建成
神经科学 > 神经解剖学和神经环路 > 皮层
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













