- EN - English
- CN - 中文
A Microplate-Based Expression Monitoring System for Arabidopsis NITRATE TRANSPORTER2.1 Using the Luciferase Reporter
基于微孔板的拟南芥硝酸盐转运蛋白2.1表达监测系统——利用荧光素酶报告基因
发布: 2024年12月05日第14卷第23期 DOI: 10.21769/BioProtoc.5127 浏览次数: 1783
评审: Vishal NehruThirupugal GovindarajanAnonymous reviewer(s)
Abstract
Gene expression analysis is a fundamental technique to elucidate the regulatory mechanisms of genes of interest or to reveal the patterns of plant response to environmental stimuli. Traditionally, gene expression analyses have required RNA extraction, followed by cDNA synthesis and qPCR analyses. However, this conventional method is costly and time-consuming, limiting the amount of data collected. The protocol outlined in this study, which utilizes a chemiluminescence system, offers a cost-effective and rapid method for assessing the expression of Arabidopsis (Arabidopsis thaliana) genes, exemplified by analyzing the nitrate-inducible expression of a major nitrate transporter gene, nitrate transporter 2.1 (NRT2.1). A reporter construct, containing the NRT2.1 promoter fused to the firefly luciferase gene, was introduced into wild-type and mutant Arabidopsis plants. Seeds obtained from the transgenic lines were grown for 3 days in 96-well microplates containing a nitrate-free nutrient solution. After 3 days, the nutrient solution was replaced with a fresh batch, which was supplemented with luciferin potassium. One hour later, nitrate was added at various concentrations, and the temporal expression pattern of NRT2.1 was analyzed by monitoring the chemiluminescence signals. This method allowed for the cost-effective, quantitative, and high-throughput analysis of NRT2.1 expression over time under the effects of various nutrient conditions and genetic backgrounds.
Key features
• Small-scale and immediate assessment of NRT2.1 promoter activity using 3-day-old Arabidopsis seedlings expressing the firefly luciferase gene under the control of the Arabidopsis NRT2.1 promoter.
• Comparison of various Arabidopsis genotypes and nutrient conditions using 96-well microplates.
• Quantitative assessment of the temporal changes in gene expression levels.
Keywords: Arabidopsis thaliana (拟南芥)Graphical overview
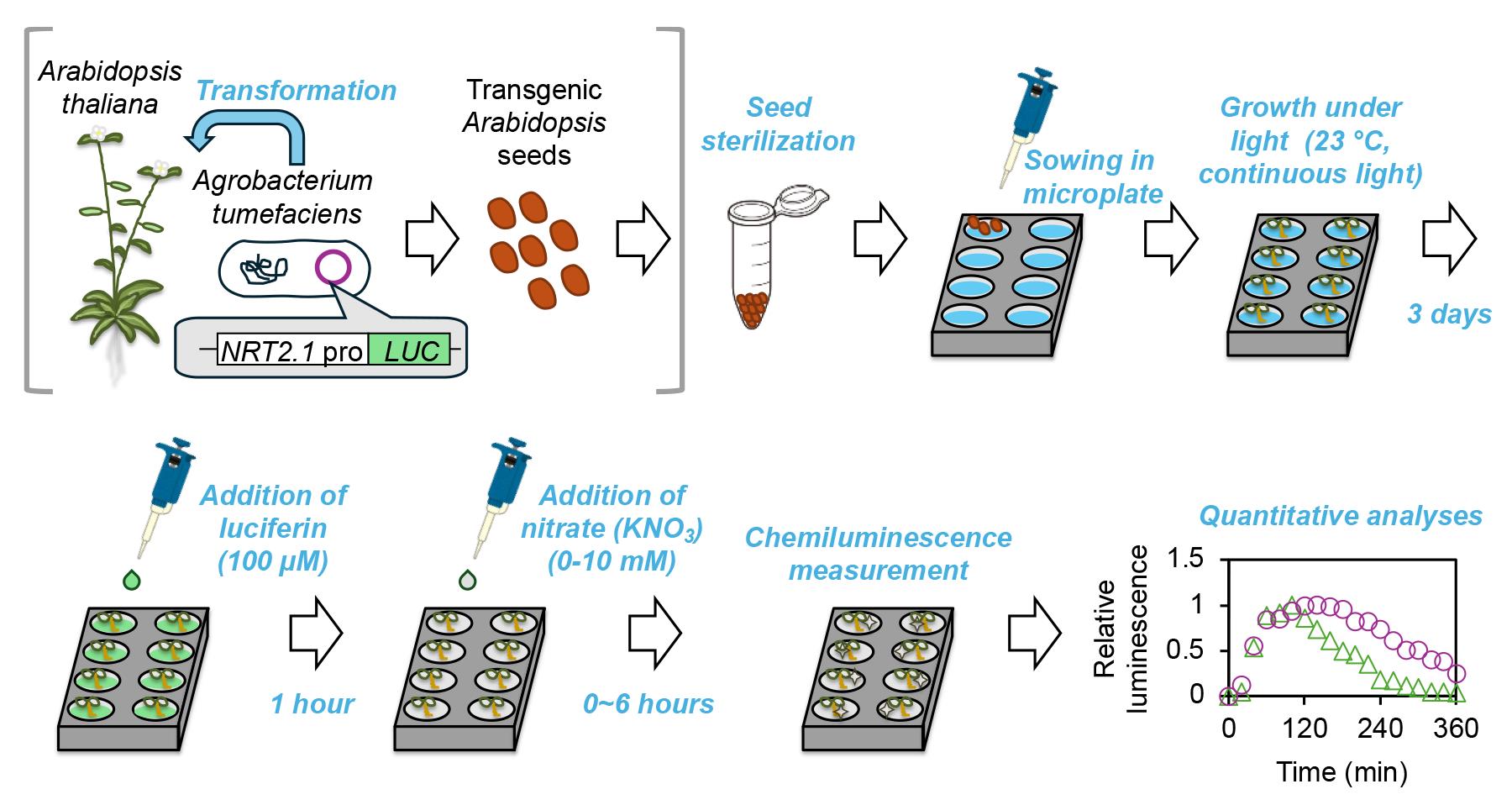
Graphical summary of the microplate-based NRT2.1 expression monitoring system in planta.
Note: The steps within gray square brackets are part of a general protocol and are not included in this manuscript.
Background
Gene expression analysis is a fundamental approach used to understand the plant response to environmental stimuli and examine the effects of gene manipulation. The current method of gene expression analysis typically involves RNA extraction, cDNA synthesis, and qPCR, which often limits the number of samples that can be handled at one time. In addition, RNA extraction and purification per se typically cost >$3 per sample, potentially hindering the analysis of a large number of samples. Consequently, many laboratories perform gene expression analyses only at a few time points, which may obscure the investigation of important genes that exhibit time-dependent expression patterns owing to the effects of multiple transcriptional regulators. In such cases, analysis of gene expression at several time points is crucial for elucidating the mechanisms employed by plants to fine-tune gene expression and for understanding the role of each transcriptional regulator in this process.
Arabidopsis nitrate transporter 2.1 gene (NRT2.1), which encodes a major high-affinity nitrate transporter required for nitrate uptake by the root [1], is an example of a gene regulated by multiple transcriptional regulators. NRT2.1 expression is under the control of multiple transcription factors with antagonistic functions, including NIN-like protein (NLP) transcription factors, which act as nitrate-activated transcriptional enhancers, and nitrate-inducible GARP-type transcriptional repressor1 (NIGT1) proteins, which act as NLP-inducible transcriptional repressors [2] (Figure 1A). NRT2.1 shows a bell-shaped expression pattern upon nitrate supply [2,3], owing to its initial NLP-induced upregulation [4], followed by NIGT1-mediated repression. To understand the contribution of different transcription factors to NRT2.1 expression, it is crucial to analyze the temporal expression pattern of NRT2.1 in different genetic backgrounds, which necessitates the analysis of NRT2.1 expression in numerous samples.
Reporter proteins, such as green fluorescent protein (GFP) and luciferase (LUC), are versatile biological tools that facilitate the analysis of protein localization or promoter activity without laborious procedures. Firefly-derived LUC gene is often used as a reporter gene to determine promoter activity in plant and animal cells because of its high sensitivity and linear range. To analyze the effects of transcription factors on target promoters, LUC is often expressed under the control of a given promoter in the analysis of plant genes in a protoplast-based assay system [5]. LUC has also been used in planta to analyze the temporal expression patterns of genes, such as those related to the circadian rhythm [6,7]. The abundance of LUC protein increases upon activation of the promoter regulating LUC expression. LUC protein converts its substrate, luciferin, to oxyluciferin with the help of a magnesium ion, which emits light. As long as the substrates (luciferin, ATP, and oxygen) are not depleted, the intensity of emitted light is assumed to be proportional to the amount of LUC protein, which enables the assessment of promoter activity (Figure 1B).
This protocol describes a detailed, step-by-step method optimized for a microplate-based LUC reporter assay system, which was used to evaluate the temporal activity of the NRT2.1 promoter. This rapid, cost-effective, and high-throughput method holds potential for analyzing the expression of various genes.
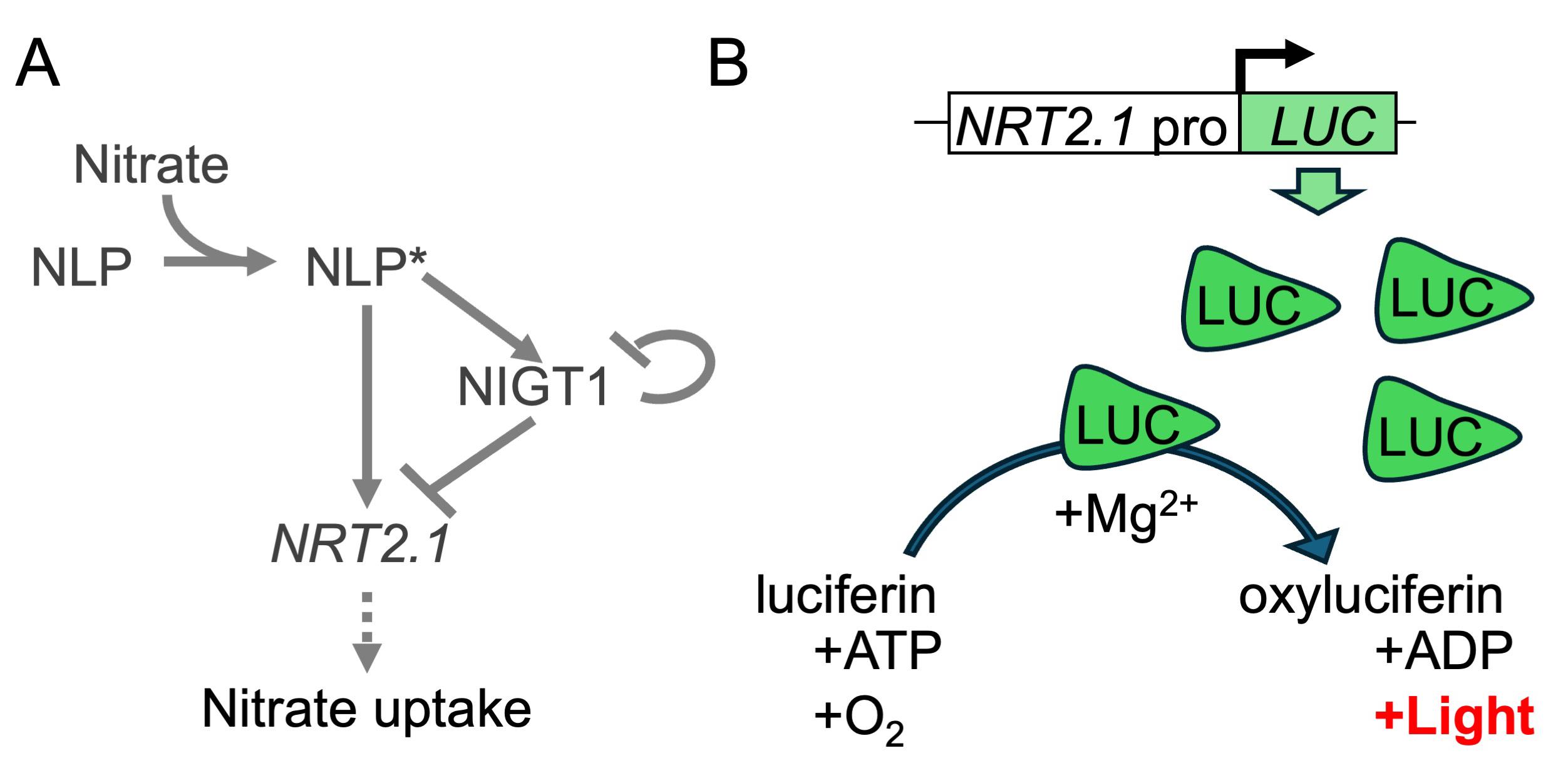
Figure 1. Regulatory mechanism of Arabidopsis NRT2.1 and the principle of luciferase (LUC) assay. (A) The known regulatory mechanism of NRT2.1 in Arabidopsis. Nitrate directly binds to and activates NLP. Activated NLP (NLP*) directly binds to the promoters of NRT2.1 and NIGT1, enhancing their expression. Subsequently, NIGT1 suppresses the expression of not only NRT2.1 but also NIGT1 genes. (B) Mode of action of LUC. The level of LUC protein increases upon the activation of NRT2.1 promoter (NRT2.1pro). LUC protein catalyzes the conversion of luciferin to an excited state (oxyluciferin) with the aid of Mg2+. Subsequently, light is emitted during the transition of oxyluciferin to the ground state.
Materials and reagents
Biological materials
Transgenic Arabidopsis thaliana seeds harboring the LUC gene under the control of NRT2.1 promoter [2]
Reagents
Sodium hypochlorite (FUJIFILM Wako Pure Chemical, catalog number: 197-02206)
D-luciferin potassium salt (FUJIFILM Wako Pure Chemical, catalog number: 120-05114)
Potassium nitrate (FUJIFILM Wako Pure Chemical, catalog number: 160-04035)
Ammonium succinate (Kanto Chemical, catalog number: 01319-30)
Murashige and Skoog salts without nitrogen, phosphorus, and iron (FUJIFILM Wako Pure Chemical, special order)
2-Morpholinoethanesulfonic acid (MES) (Dojindo, catalog number: 343-01626)
Potassium hydroxide (FUJIFILM Wako Pure Chemical, catalog number: 168-21815)
Potassium dihydrogen phosphate (FUJIFILM Wako Pure Chemical, catalog number: 169-02425)
Iron (II) sulfate heptahydrate (FUJIFILM Wako Pure Chemical, catalog number: 094-01082)
Sucrose (FUJIFILM Wako Pure Chemical, catalog number: 196-00015)
Solutions
MES buffer (see Recipes)
10 mM iron (II) sulfate solution (see Recipes)
1 M potassium dihydrogen phosphate solution (see Recipes)
100 mM ammonium succinate solution (see Recipes)
1× Murashige and Skoog solution (N-, P-, Fe-free) (see Recipes)
0.1× N-free MS medium (see Recipes)
1 M potassium nitrate stock solution (see Recipes)
10 mM D-luciferin potassium stock solution (see Recipes)
Recipes
170 mM MES buffer
Note: Adjust the pH of 170 mM MES buffer to 5.7 using potassium hydroxide solution. Filter-sterilize the buffer and store at room temperature.
Reagent Final concentration Quantity or Volume 2-Morpholinoethanesulfonic acid 170 mM 3.6 g Potassium hydroxide n/a n/a Ultrapure water (Milli-Q) n/a Up to 100 mL Total n/a 100 mL 10 mM iron (II) sulfate stock solution (10 mL)
Note: Filter the solution, aliquot into 1.5 mL tubes, and store at -20 °C to prevent precipitation.
Reagent Final concentration Quantity or Volume Iron (II) sulfate heptahydrate 10 mM 27.8 mg Ultrapure water (Milli-Q) n/a Up to 10 mL Total n/a 10 mL 1 M potassium dihydrogen phosphate stock solution (10 mL)
Note: Autoclave or filter-sterilize the solution and store at room temperature.
Reagent Final concentration Quantity or Volume Potassium dihydrogen phosphate 1 M 1.36 g Ultrapure water (Milli-Q) n/a Up to 10 mL Total n/a 10 mL 100 mM ammonium succinate stock solution (10 mL)
Note: Autoclave or filter-sterilize the solution and store at room temperature.
Reagent Final concentration Quantity or Volume Ammonium succinate 100 mM 0.15 g Ultrapure water (Milli-Q) n/a Up to 10 mL Total n/a 10 mL 1× Murashige and Skoog medium (without N, P, and Fe) (1 L)
Note: To conduct studies involving nutrient-deficiency treatments, make a specialized Murashige and Skoog plant salt mixture lacking KNO3, NH4NO3, KH2PO4, and FeSO4, and store the solution at 4 °C.
Reagent Final concentration Quantity or Volume Murashige and Skoog plant salt mixture (N-, P-, and Fe-free) 1× 1 bag Ultrapure water (Milli-Q) n/a Up to 1 L Total n/a 1 L 0.1× N-free MS medium + 500 µM ammonium succinate (100 mL)
Note: Mix all components, except iron (II) sulfate solution, and autoclave. After autoclaving, add the iron (II) sulfate solution and mix thoroughly. Store the solution at 4 °C.
Reagent Final concentration Quantity or Volume 1× Murashige and Skoog medium (without N, P, and Fe) 0.1× 10 mL Sucrose 0.5% (w/v) 0.5 g 100 mM ammonium succinate stock solution 500 µM 0.5 mL 1 M potassium dihydrogen phosphate stock solution 125 µM 12.5 µL 170 mM MES stock solution 3.5 mM 2.1 mL 10 mM iron (II) sulfate stock solution (add after autoclave) 10 µM 100 µL Ultrapure water (Milli-Q) n/a Up to 100 mL Total n/a 100 mL 1 M potassium nitrate stock solution (10 mL)
Note: Autoclave or filter-sterilize the solution and store at room temperature.
Reagent Final concentration Quantity or Volume Potassium nitrate 1 M 1.0 g Ultrapure water (Milli-Q) n/a Up to 10 mL Total n/a 10 mL 10 mM luciferin potassium stock solution (10 mL)
Note: Aliquot the solution into 1.5 mL tubes and store at -80 °C.
Reagent Final concentration Quantity or Volume Luciferin potassium 10 mM 31.8 mg Ultrapure water (Milli-Q) n/a Up to 10 mL Total n/a 10 mL
Laboratory supplies
Black 96-well microplates (Greiner, F-bottom, catalog number: 655077)
Transparent lids for 96-well plates (e.g., TPP, catalog number: 92696)
1.5 mL tubes (e.g., Watson, catalog number: 131-415C)
200 µL pipette tips (e.g., Watson, catalog number: 110-705C)
Surgical tape (e.g., 3M Company, catalog number: 1530-0)
Beaker
Equipment
Microplate reader (Tecan, model: Infinite M1000)
Electronic balance
Autoclave
Tabletop centrifuge
pH meter
Magnetic stirrer
Clean bench
Growth chamber (equipped with LED lights)
Pipettes (200 µL, single- and 8-channel)
Software and datasets
Magellan software (Tecan)
Microsoft Excel (Microsoft)
Procedure
文章信息
稿件历史记录
提交日期: Jul 11, 2024
接收日期: Oct 7, 2024
在线发布日期: Oct 22, 2024
出版日期: Dec 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ueda, Y. and Yanagisawa, S. (2024). A Microplate-Based Expression Monitoring System for Arabidopsis NITRATE TRANSPORTER2.1 Using the Luciferase Reporter. Bio-protocol 14(23): e5127. DOI: 10.21769/BioProtoc.5127.
- Ueda, Y. and Yanagisawa, S. (2023). Transcription factor module NLP–NIGT1 fine-tunes NITRATE TRANSPORTER2.1 expression. Plant Physiol. 193(4): 2865–2879.
分类
植物科学 > 植物分子生物学 > DNA > 基因表达
分子生物学 > DNA > 基因表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link