- EN - English
- CN - 中文
Generation of Zebrafish Maternal Mutants via Oocyte-Specific Knockout System
基于卵母细胞特异性敲除系统生成斑马鱼母源突变体
(*contributed equally to this work) 发布: 2024年11月05日第14卷第21期 DOI: 10.21769/BioProtoc.5092 浏览次数: 2213
评审: Anonymous reviewer(s)
Abstract
Maternal mRNAs and proteins are produced during oogenesis by more than 60% of zebrafish genes. They are indispensable for fertilization and early embryogenesis. Generation and analysis of the maternal mutant is the most direct way to characterize the maternal function of the specific gene. However, due to the lethality of zygotic mutants, the maternal function of most genes in zebrafish remains elusive. Several methods have been developed to circumvent this obstacle, including mRNA rescue, germ-line replacement, oocyte microinjection in situ, mosaic mutation, and bacterial artificial chromosome (BAC)-mediated conditional rescue. Here, we provide an alternative approach to generate zebrafish maternal mutants rapidly and efficiently by introducing four tandem sgRNA expression cassettes into Tg(zpc:zcas9) embryos. This method is more technically feasible and cost- and time-effective than other established methods.
Key features
• This protocol can circumvent the lethality or infertility of the zygotic mutants to obtain maternal mutants of the target gene.
• This protocol is time-saving (one fish generation).
• Using this protocol, double-gene maternal mutants can be obtained in a single generation.
• Stable lines can be established to continuously produce maternal mutant embryos for the gene of interest.
Keywords: Zebrafish (斑马鱼)Graphical overview
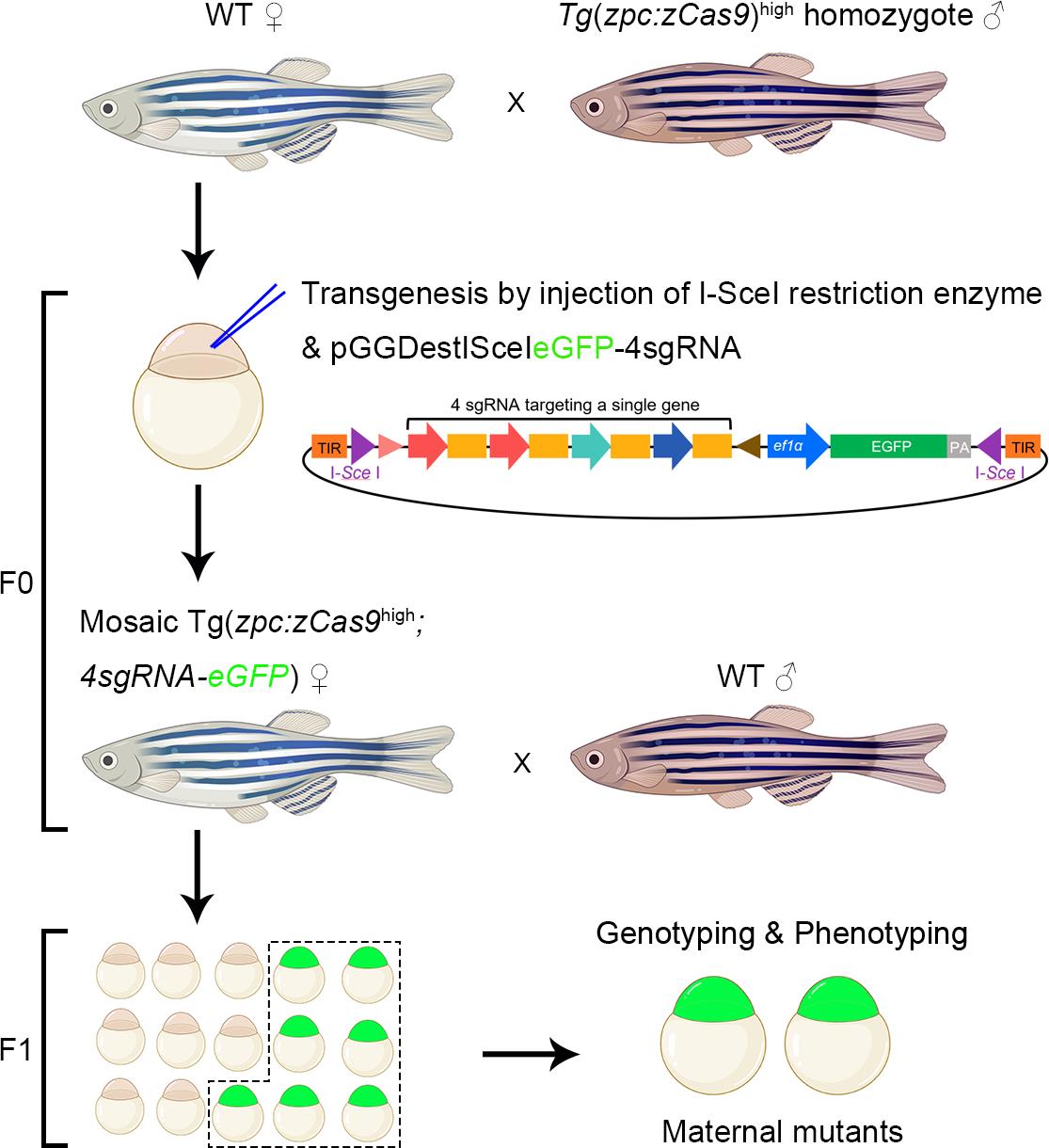
Background
Maternal factors are mRNAs and proteins deposited in the oocyte. They play vital roles in oocyte maturation, fertilization, and blastocyst development. Approximately 66% of zebrafish genes are expressed maternally [1]. As zygotic genome activation (ZGA) starts at around the 1k-cell stage, maternal factors are dominant to function in the developmental events before this time point. In addition, even after zygotic genome activation (ZGA), maternal factors still function in later embryogenesis, including axis formation, germ layers differentiation, morphogenesis, etc. [2–4].
Generation of the corresponding maternal mutant is the most straightforward way to address the maternal function of a gene in zebrafish. The genotype of primary oocyte is the same as that of somatic cells. Hence, viable and fertile female zygotic mutants are indispensable for giving birth to maternal mutants or maternal and zygotic mutants. However, zygotic mutants usually cannot survive to adulthood or spawn because of the crucial and uncompensable functions of the mutated genes [5–7]. This obstacle represents the major bottleneck to studying maternal factors. Several methods have been developed to bypass this technical hurdle. The first is mRNA rescue. Through microinjection of in vitro–synthesized wild-type mRNA, the zygotic mutant can be rescued to live through a critical period, in which the zygotic products contribute to normal development [4]. Considering that the ectopic expression may disrupt normal embryogenesis and transient expression of mRNA cannot support long-term gene function, this method is not applicable to most genes. In 2002, Ciruna et al. developed the germ-line replacement technology. They injected dead end1 morpholino into wild-type embryos to block primordial germ cell development and then transplanted germ cells from zygotic mutants into these primordial germ cell (PGC)-free hosts. This ensured that the germ lines of the host were entirely replaced by donor PGCs. Upon maturation, all the embryos laid by the female chimera were maternal mutants [8]. Transplantation of PGCs is technically demanding and the chimeric embryos tend to develop into males because of the limited number of transplanted PGCs. Wu et al. established a surgery-based method [9]. Following the opening of the female fish abdomen, the fluorescent lineage tracer and the morpholino targeting the specific gene were co-injected into the stage- oocyte. After recovery, the laid fluorescent embryos were the desired maternal morphants [9]. However, the intricate operative skills required for the application of this method are significantly challenging. In 2018, Xing et al. generated mosaic mutants containing homozygous mutant cells through secondary genome editing in heterozygotes [2]. This approach was designed to circumvent the lethality associated with double zygotic mutants of dvl2 and dvl3a. Some of these homozygous mutant cells were incorporated into the germline, eventually resulting in maternal mutants after fertilization. However, this method is inefficient and time-consuming, requiring at least three generations. In addition to the methods mentioned above, the bacterial artificial chromosome-rescue-based knockout (BACK) is another method to bypass the zygotic lethality and obtain maternal mutants [10]. To obtain such mutants, first, the authors generated mutants of the target gene through nuclease-mediated genome editing. Second, they rescued the zygotic mutants by introducing a bacterial artificial chromosome (BAC) containing the target gene expression cassette flanked by loxP sites via Tol2-mediated transgenesis. Third, they crossed these fish with a germline-specific Cre line. This approach ultimately allowed them to obtain maternal mutants of the target gene. However, this strategy is time-consuming and labor-intensive, as it involves gene knock-out, transgenesis, and crossing with the Cre line.
Recently, we have developed a new strategy to generate single or double-gene maternal mutants through transgenic expression of Cas9 and sgRNA in oocytes [11,12]. Through I-Sce I-mediated transgenesis, we introduced an eGFP reporter and multiple tandem sgRNA expression cassettes into Tg(zpc:zcas9) transgenic fish, where Cas9 is specifically expressed in oocytes. This approach enables genome editing during the early stages of oogenesis. As a result, some of the GFP-positive embryos will have their maternal products completely ablated, thereby becoming maternal mutants. Consequently, we can analyze their phenotypes after identifying them through genotyping. Compared to other methods, it has several advantages. First, it is technically feasible. The main techniques are plasmid construction and transgenesis, which are conventional in most zebrafish labs. Second, it is time saving, taking one generation (2–3 months) for either single or double-genes maternal knockout. Third, the efficiency is generally stable despite individual variation. Through generating maternal mutants of nanog, ctnnb2, rbm24a, dvl2, and dvl3a, we find that the average ratio of the maternal mutant for a single gene is approximately 25% and the maximum efficiency can reach 63.3%. For every single founder fish, the ratio of maternal mutants remains stable in each spawning, so that we can obtain maternal mutants repeatedly once we get a founder. Finally, this approach can be utilized to generate double-gene maternal mutants, which takes nearly the same time.
Materials and reagents
Biological materials
Zebrafish lines:
Wild-type AB zebrafish line was obtained from the China zebrafish resource center
Tg(zpc:zcas9) transgenic line was reported by Ming Shao’s lab and is available upon request [13]
Reagents
NaCl (Solarbio, catalog number: S8210)
KCl (Solarbio, catalog number: P9921)
CaCl2·2H2O (Solarbio, catalog number: C8370)
HEPES (Sigma-Aldrich, catalog number: H3375)
2× Taq Master Mix (Dye Plus) (Vazyme, catalog number: P112-01)
AxyPrep PCR Clean-Up Kit (Axygen, catalog number: AP-PCR-250)
T7 RNA polymerase, 5× transcription buffer (Thermo Scientific, catalog number: EP0111)
ATP/CTP/GTP/UTP, 10 mM each (Thermo Scientific, catalog number: R0481ribo); dilute the 100 mM solution with DNase/RNase-free H2O
RiboLock RNase inhibitor (Thermo Scientific, catalog number: EO0381)
TURBO DNase (Invitrogen, catalog number: AM2238)
Ammonium acetate, 5 M, RNase-free (Thermo Scientific, catalog number: AM9070G)
Phenol:chloroform:isoamyl alcohol 25:24:1, pH 5.2 (Thermo Scientific, catalog number: J62336.AE)
Cas9 protein: GenCrispr NLS-Cas9-NLS nuclease (GenScript, catalog number: Z03389-50)
AxyPrep Plasmid Miniprep Kit (Axygen, catalog number: AP-MN-P-250)
Acc65I (Asp718I) (Thermo Scientific, catalog number: ER0901)
Gibson assembly kit: Hieff Clone Plus One Step Cloning Kit (Yeasen, catalog number: 10911ES20)
CutSmart buffer (NEB, catalog number: B60004)
Agarose (BIOWEST, catalog number: 111860)
Glass capillaries (World Precision Instruments, catalog number: TWF-100F-4)
100× penicillin-streptomycin solution (Gibco, catalog number: 15140122)
cDNA synthesis kit (Transgene, catalog number: AT301)
pBackZero-T vector (Takara, catalog number: 3275)
Primers:
Universal primer:
5'-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGC
TATTTCTAGCTCTAAAAC-3'
Dest forward primer: 5'-TTCTTGTTTAAGCTTTTAATCTCAAAAAAC-3'
Dest reverse primer. 5'-GGCTGTTTACATCTGATAGTGG-3'
Ligation forward primer: 5'-gagtcggtgctttttttaaacctggTTCTTGTTTAAGCTTTTAATCTCAAAAAAC-3'
Ligation reverse primer: 5'-ATCCTGCACTGAATGCAC-3'
bmp2b sgRNA forward primer: 5'- taatacgactcactataGGGAGGCTGAGAGCAACCGGgttttagagctagaa-3'
M13 forward: 5'-GTAAAACGACGGCCAGT-3'
The plasmid system is available upon request to the authors Chong Zhang and Ming Shao
TRIzol (Thermo Scientific, catalog number: 15596026)
PBST (Solarbio, catalog number: P1031)
Chloroform (Sinopharm Chemical Reagent, catalog number: 10006862)
Isopropanol (Sinopharm Chemical Reagent, catalog number: 40064360)
Glycogen (Thermo Scientific, catalog number: R0561)
Solutions
Ringer's buffer (see Recipes)
Recipes
Ringer's buffer
Reagent Final concentration Quantity NaCl 116 mM 6.779 g KCl 2.9 mM 0.216 g CaCl2·2H2O 1.8 mM 0.265 g HEPES 5 mM 1.192 g H2O n/a 1,000 mL Total n/a 1,000 mL Dissolve all ingredients in 900 mL of deionized H2O. Adjust pH to 7.2 using NaOH. After being autoclaved, it can be long-term stored at room temperature.
Equipment
VeritiTM Dx 96-well thermal cycler (Thermo Fisher, catalog number: 4452300)
NanoDrop 2000 spectrophotometer (Thermo Scientific, model: ND-2000)
Block heater (Yiheng, model: TU-100C)
Electrophoresis system (Bio-Rad, catalog number: 1640302)
Centrifuge (Eppendorf, model: 5424R)
Pico-injector (Warner Instrument, model: PLI-100A)
Puller (NARISHIGE, model: PC-100)
Tweezer (WPI, catalog number: 500341)
Microforge (NARISHIGE, model: MF2)
Microinjector (Harvard Apparatus, model: PLI-100A)
Software and datasets
CRISPRScan, http://www.crisprscan.org/, for designing sgRNAs with high on-target activity and minimal off-target effects
Synthego, https://ice.synthego.com/#/, for analyzing sequencing results and assessing CRISPR editing efficiency
Procedure
文章信息
稿件历史记录
提交日期: Jun 10, 2024
接收日期: Aug 28, 2024
在线发布日期: Oct 13, 2024
出版日期: Nov 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Zhang, C., Wei, W., Lu, T., Zhang, Y., Li, J., Wang, J., Chen, A., Wen, F. and Shao, M. (2024). Generation of Zebrafish Maternal Mutants via Oocyte-Specific Knockout System. Bio-protocol 14(21): e5092. DOI: 10.21769/BioProtoc.5092.
分类
发育生物学 > 基因组编辑
生物科学 > 生物技术 > CRISPR/Cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













