- EN - English
- CN - 中文
Metabolic RNA Labeling and Translating Ribosome Affinity Purification for Measurement of Nascent RNA Translation
过代谢RNA标记和翻译核糖体亲和纯化测量新生RNA的翻译
发布: 2024年10月20日第14卷第20期 DOI: 10.21769/BioProtoc.5091 浏览次数: 2521
评审: Alessandro DidonnaHemant Kumar PrajapatiWeidong An
Abstract
Regulation of gene expression in response to various biological processes, including extracellular stimulation and environmental adaptation, requires nascent mRNA synthesis and translation. Simultaneous analysis of the coordinated regulation of dynamic mRNA synthesis and translation using the same experiment remains a major challenge in the field. Here, we describe a step-by-step protocol for the simultaneous measurement of transcription of nascent mRNA and its translation at the gene level during the acute unfolded protein response (UPR) in HEK293 cells by combining 4-thiouridine metabolic mRNA labeling with translational ribosome affinity purification (TRAP) using a monoclonal antibody against evolutionarily conserved ribosomal P-stalk proteins (P-TRAP). Since P-TRAP captures full-length RNAs bound to ribosomes, it is compatible with 3' mRNA-seq, which analyzes the uridine-rich 3' UTRs of polyadenylated RNAs, allowing robust quantification of T>C conversions. Our nascent P-TRAP (nP-TRAP) method, in which P-TRAP is combined with metabolic mRNA labeling, can serve as a simple and powerful tool to analyze the coordinated regulation of transcription and translation of individual genes in cultured cells.
Key features
• Simple and retriable analysis of nascent mRNA synthesis and its translation in cultured cells
• Combination of 4-thiouridine metabolic RNA labeling with translating ribosome affinity purification (TRAP)
• Ribosomal P-stalk-mediated TRAP (P-TRAP) allows single-step and efficient purification of non-tagged ribosomes and translated mRNAs
Keywords: Translating ribosome affinity purification (翻译核糖体亲和纯化)Graphical overview
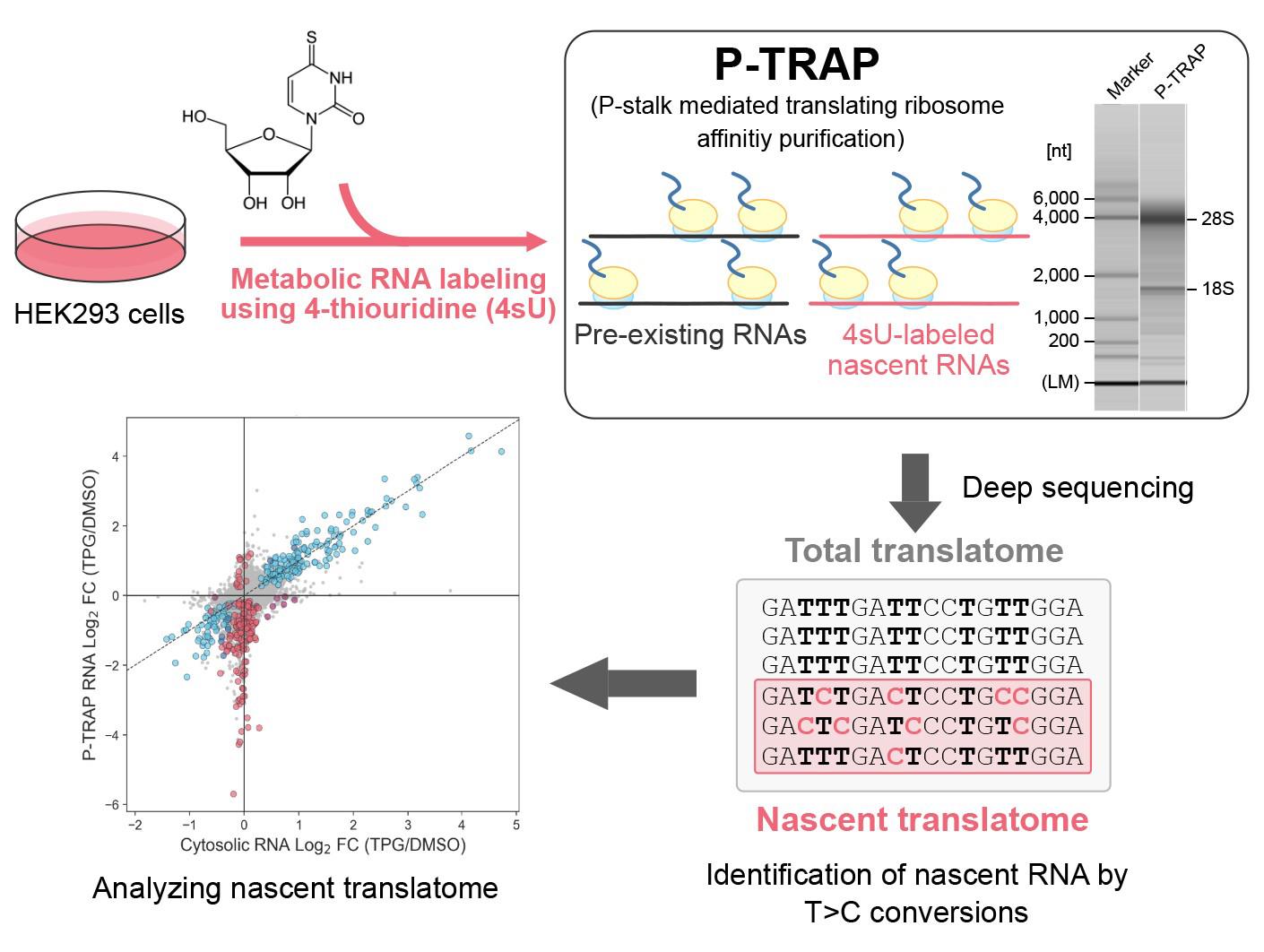
Background
Regulation of gene expression plays a central role in many biological processes. The final output of a protein-coding gene, the amount of protein, is determined by several regulation processes: transcription and degradation of mRNA, translation of mRNA, and degradation of protein.
Metabolic RNA labeling techniques have been developed to analyze the dynamic transcription of nascent mRNA and its degradation. One of these labeling techniques is a thiol (SH)-linked alkylation for the metabolic sequencing of RNA (SLAMseq), in which RNAs are labeled with the uridine analog 4-thiouridine (4sU) [1]. 4sU is incorporated into nascent RNAs by RNA polymerase II and then alkylated during cDNA library preparation, allowing bioinformatic analysis to detect the specific T-to-C (T>C) conversion at the 4sU incorporation site in 4sU-labeled RNA. By focusing on sequence reads containing T>C conversions, the accumulation rate of nascent transcribed RNA after the addition of 4sU (progressive labeling technique) or the degradation rate of pre-4sU-labeled RNA after the addition of an excess amount of uridine (pulse-chase technique) can be analyzed [2,3].
In addition to mRNA transcription and its degradation, mRNA translation significantly influences the level of gene expression [4]. Currently, to analyze the level of translation, three different methods are employed utilizing next-generation sequencing techniques: polysome profiling, translating ribosome affinity purification (TRAP), and ribosome profiling (Ribo-seq). Polysome profiling fractionates full-length ribosome-bound RNAs using sucrose density gradient ultracentrifugation. TRAP also fractionates full-length ribosome-bound RNAs, but it relies on the immunoprecipitation of ribosomes. Ribo-seq fractionates ribosome-protected RNA fragments (RPFs) after sucrose density gradient via ultracentrifugation and RNase digestion. Each method has advantages and disadvantages and is used according to the experimental design and objectives (reviewed in [5]).
The most dominant regulation process that determines the protein level is different for each gene. Therefore, the simultaneous measurement of these regulation processes in a single experiment is advantageous to understand the regulatory mechanism of gene expression, which is still a major challenge in this field. Kawata et al. simultaneously evaluated the synthesis rate of nascent mRNA and the degradation rate of pre-existing mRNA by using two different metabolic labeling analogs, i.e., 5'-bromo-uridine (BrU) and 4sU [6]. Statistical approaches have also been developed to estimate both the synthesis rate of nascent mRNA and the degradation rate of pre-existing mRNA using only 4sU labeling [2,7]. Recently, Schott et al. developed nascent Ribo-seq (nRibo-seq), a combination of 4sU metabolic RNA labeling and Ribo-seq, which allows simultaneous measurement of nascent mRNA synthesis and its translation at the level of bulk or specific RNA groups [8]. Although nRibo-seq is a pioneering approach for measuring mRNA transcription and translation, Ribo-seq deals with the short length of RPFs, making the reliable quantification of 4sU incorporation for individual genes difficult [8].
Here, we describe a step-by-step protocol for the simultaneous measurement of transcription of nascent mRNA and its translation at the gene level in the acute unfolded protein response (UPR), in which transcription and translation are dynamically reprogrammed to reduce unfolded and misfolded proteins in the endoplasmic reticulum and restore protein homeostasis [9] in HEK293 cells by combining 4sU metabolic mRNA labeling with TRAP (originally reported in [10]). The use of a monoclonal antibody against the evolutionarily conserved ribosomal proteins P0, P1, and P2 (P-stalk) is responsible for the highly efficient purification of endogenous translating ribosomes without an affinity tag (e.g., FLAG-tag, His-tag, and SBP-tag), shortening the experimental period (termed as the P-TRAP method). Combining this P-TRAP with 4sU metabolic mRNA labeling and 3' mRNA-Seq (which analyzes the uridine-rich 3' UTR of polyadenylated RNAs) results in robust quantification of T>C conversion and reliable analysis of nascent mRNA transcription as well as its translation at the individual gene level. If the sensitivity of nascent RNA detection is sufficient, subtracting T>C reads from total reads will yield non-T>C reads that are mostly derived from pre-existing RNAs, contributing to further analysis of translation in not only nascent but also pre-existing RNAs. This simple and versatile nascent P-TRAP (nP-TRAP) method could enhance our understanding of the complex regulation of gene expression in eukaryotes.
Materials and reagents
Biological materials
Flp-InTM T-REXTM 293 cell line (Thermo Fisher Scientific, catalog number: R78007)
DMEM (high glucose) with L-Glutamine, phenol red and sodium pyruvate (FUJIFILM Wako, catalog number: 043-30085)
Penicillin-streptomycin mixed solution (Nacalai Tesque, catalog number: 09367-34)
Fetal bovine serum (FBS) (Cell Culture Bioscience, catalog number: 171012)
D-PBS(-) without Ca and Mg, liquid (Nacalai Tesque, catalog number: 14249-24)
Nuclease-free water (not DEPC-treated) (Thermo Fisher Scientific, catalog number: AM9937)
HEPES (Merck, catalog number: H3375-500G)
Tris (Nacalai Tesque, catalog number: 035401-25)
HCl (FUJIFILM, catalog number: 080-01066)
Glycine (FUJIFILM, catalog number: 077-00735)
MgCl2 (Nacalai Tesque, catalog number: 20908-65)
NaCl (FUJIFILM Wako, catalog number: 191-01665)
NaH2PO4·H2O (Nacalai Tesque, catalog number: 31719-05)
Na2HPO4·12H2O (FUJIFILM Wako, catalog number: 196-02835)
DTT (Roche, catalog number: 10708984001)
Triton X-100 (Nacalai Tesque, catalog number: 35501-15)
Tween 20 (FUJIFILM, catalog number: 167-11515)
SDS (SERVA, catalog number: 20765.02)
DMSO (FUJIFILM Wako, catalog number: 043-07211)
Glycerol (FUJIFILM Wako, catalog number: 075-00616)
Bromophenol blue (FUJIFILM Wako, catalog number: 021-02911)
BSA (Nacalai Tesque, catalog number: 01860-07)
Protease inhibitor cocktail for use with mammalian cell and tissue extracts (Nacalai Tesque, catalog number: 25955-11)
Phosphatase inhibitor cocktail (EDTA-free) (Nacalai Tesque, catalog number: 07575-51)
4-thiouridine (4sU) (LKT Laboratory, catalog number: T2933)
Thapsigargin (Cayman Chemical, catalog number: 10522)
Cycloheximide (Nacalai Tesque, catalog number: 06741-91)
RNase A (100 mg/mL) (NIPPON GENE, catalog number: 318-06391)
RNasin® Plus ribonuclease inhibitor (Promega, catalog number: N2611)
DynabeadsTM protein G for immunoprecipitation (Thermo Fisher Scientific, catalog number: 10004D)
Anti-ribosomal proteins P0/P1/P2 mAb [9D5] (MBL, catalog number: RN004M)
Mouse IgG2a isotype control (Proteintech, catalog number: 65208-1-Ig)
Antibodies for western blotting:
Anti-ribosomal protein uL3 (GeneTex, catalog number: GTX114725)
Anti-ribosomal protein uS2 (GeneTex, catalog number: GTX114734)
Anti-PABP4 (BETHYL, catalog number: A301-467A), primary antibody
Anti-CBP80 (BETHYL, catalog number: A301-793A), primary antibody
Anti-eIF4A3 [11], primary antibody
Anti-GAPDH mAb-HRP-DirecT (MBL, catalog number: M171-7)
Anti-rabbit IgG, HRP-linked antibody (Cell Signaling Technology, catalog number: 7074S)
Anti-mouse IgG, HRP-linked antibody (Cell Signaling Technology, catalog number: 7076S)
ProClinTM 950 (Merck, catalog number: 46878-U)
e-PAGEL 5%–20% 18-well precast mini gel (ATTO, catalog number: E-R520L)
Pre-stained XL-Ladder Broad (APRO, catalog number: SP-2110)
EzFastBlot Fast western blotting transfer buffer (ATTO, catalog number: AE-1465)
ECL selectTM western blotting detection reagent (Cytiva, catalog number: RPN2235)
ImmunoStar® LD (FUJIFILM, catalog number: 296-69901)
ISOGEN II (NIPPON GENE, catalog number: 311-07361)
2-Propanol (Nacalai Tesque, catalog number: 29113-95)
Ethanol (99.5) (Nacalai Tesque, catalog number: 14713-95)
Iodoacetamide, No-WeighTM format (Thermo Fisher Scientific, catalog number: A39271)
Sodium acetate nuclease and protease tested (Nacalai Tesque, catalog number: 31137-25)
3 M NaOAc pH 5.2 (FUJIFILM Wako, catalog number: 316-90081)
10 N NaOH (Nacalai Tesque, catalog number: 94611-45)
Glycogen solution (20 mg/mL) from Oyster, nuclease tested (Nacalai Tesque, catalog number: 17110-11)
TE buffer pH 8.0 (Nacalai Tesque, catalog number: 06890-54)
RNA 1000 reagent kit for MultiNA (SHIMADZU, catalog number: 292-27913-91)
SYBRTM Green II RNA gel stain, 10,000× concentrate in DMSO (Invitrogen, catalog number: S7568)
RNA 6000 ladder (Invitrogen, catalog number: AM7152)
Formamide (Nacalai Tesque, catalog number: 16229-95)
QuantSeq 3' mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen, catalog number: 192.14)
Note: We used the currently unavailable catalog number 015.24. One may use 191.24 as an alternative.
Qubit® dsDNA HS Assay Kits (molecular probes life technologies, catalog number: Q32854)
20× TBS stock solution
1 M Tris-HCl pH 7.5 stock solution
1 M MgCl2 stock solution
5 M NaCl stock solution
1 M DTT stock solution
1% SDS stock solution
Tris-glycine SDS running buffer (see Recipes)
1 M HEPES-NaOH pH 7.5 stock solution (see Recipes)
100 mg/mL cycloheximide (see Recipes)
1% Triton X-100 lysis buffer (see Recipes)
Wash buffer (see Recipes)
SDS buffer (see Recipes)
6× SDS-dye (see Recipes)
TBST (see Recipes)
1% (w/v) BSA/TBST (see Recipes)
200 mM 4-thiouridine (4sU) (see Recipes)
1 mM Thapsigargin (TPG)
1 M NaH2PO4 (see Recipes)
1 M Na2HPO4 (see Recipes)
0.5 M NaPO4 pH 8.0 (see Recipes)
100 mM IAA (see Recipes)
Tris-glycine SDS running buffer (2,000 mL)
Reagent Final concentration Quantity or Volume Tris 25 mM 6 g Glycine 192 mM 28.8 g SDS 0.1% (w/v) 2 g Deionized water n/a Up to 2,000 mL Note: Store at room temperature (RT).
1 M HEPES-NaOH pH 7.5 (1,000 mL)
Reagent Final concentration Quantity or Volume HEPES 1 M 238.3 g 10 N NaOH n/a To pH 7.5 Deionized water n/a Up to 1,000 mL Notes:
Prepare ~800 mL of deionized water in a suitable container. Add 238.3 g of HEPES to the solution. Adjust solution to the desired pH using 10 N NaOH. Add deionized water until the volume is 1,000 mL.
Store at 4 °C.
100 mg/mL cycloheximide (0.1 mL)
Reagent Final concentration Quantity or Volume Cycloheximide 100 mg/mL 10 mg DMSO n/a 0.1 mL Note: Prepare before use.
1% Triton X-100 lysis buffer (10 mL)
Reagent Final concentration Quantity or Volume 1 M HEPES-NaOH pH 7.5 20 mM 0.2 mL 1 M MgCl2 2.5 mM 0.025 mL 5 M NaCl 150 mM 0.3 mL Triton X-100 1% (v/v) 0.1 mL 1 M DTT 0.5 mM 5 μL Cycloheximide (100 mg/mL) 100 μg/mL 10 μL Protease inhibitor cocktail (100×) 1× 0.1 mL Phosphatase inhibitor cocktail (100×) 1× 0.1 mL Nuclease-free water n/a Up to 10 mL Notes:
Store at RT.
Add DTT, cycloheximide, protease inhibitor cocktail, and phosphatase inhibitor cocktail before use.
Wash buffer (50 mL)
Reagent Final concentration Quantity or Volume 1 M HEPES-NaOH pH 7.5 20 mM 1 mL 1 M MgCl2 2.5 mM 0.125 mL 5 M NaCl 150 mM 1.5 mL Tween 20 0.025% (v/v) 12.5 μL Nuclease-free water n/a 47.375 mL Note: Store at RT.
SDS buffer (10 mL)
Reagent Final concentration Quantity or Volume 1 M HEPES-NaOH pH 7.5 20 mM 1 mL 1% SDS 1% 1 mL Nuclease-free water n/a 8 mL Note: Store at RT.
6× SDS-dye (10 mL)
Reagent Final concentration Quantity or Volume 1 M Tris-HCl pH 7.5 0.3 M 3 mL SDS 9% (w/v) 0.9 g Glycerol 30% (v/v) 3 mL 1 M DTT 0.6 M 6 mL Bromophenol blue 0.1% (w/v) 10 mg Nuclease-free water n/a Up to 10 mL Note: Store at -20 °C.
TBST (2,000 mL)
Reagent Final concentration Quantity or Volume 20× TBS 1× 100 mL Tween 20 0.1% (v/v) 2 mL Deionized water n/a Up to 2,000 mL Note: Store at RT.
1% BSA/TBST (50 mL)
Reagent Final concentration Quantity or Volume 1× TBST 1× 50 mL BSA 1% (w/v) 0.5 g Note: Prepare before use.
200 mM 4-thiouridine (0.5 mL)
Reagent Final concentration Quantity or Volume 4-thiouridine (MW 260.27) 200 mM 26 mg Nuclease-free water n/a 0.5 mL Note: Store at -20 °C and protected from light.
1 mM Thapsigargin (1.5 mL)
Reagent Final concentration Quantity or Volume Thapsigargin (MW 650.8) 1 mM 1 mg DMSO n/a 1.536 mL Note: Dispense 20 μL each and store at -80 °C.
1 M NaH2PO4 (10 mL)
Reagent Final concentration Quantity or Volume NaH2PO4·H2O 1 M 1.38 g Nuclease-free water n/a Up to 10 mL Note: Store at RT.
1 M Na2HPO4 (10 mL)
Reagent Final concentration Quantity or Volume Na2HPO4·12H2O 1 M 3.58 g Nuclease-free water n/a Up to 10 mL Note: Store at RT.
0.5 M NaPO4 pH 8.0 (10 mL)
Reagent Final concentration Quantity or Volume 1 M NaH2PO4 0.034 M 0.34 mL 1 M Na2HPO4 0.466 M 4.66 mL Nuclease-free water n/a 5 mL Note: Store at RT after 0.22 μm filtration.
100 mM IAA (0.5 mL)
Reagent Final concentration Quantity or Volume Iodoacetamide, No-WeighTM 100 mM 9.3 mg (1 vial) EtOH n/a 0.5 mL Note: Prepare before use.
Laboratory supplies
Cell culture 6-well plate (SPL Life science, catalog number: 30006)
Tissue culture dish Φ96 × 21 mm (10 cm) (TPP, catalog number: 93100)
Falcon® 5 mL serological pipette (CORNING, catalog number: 357543)
Falcon® 10 mL serological pipette (CORNING, catalog number: 357551)
FastGeneTM centrifuge tubes 15 mL (NIPPON Genetics, catalog number: FG400)
SuperClear® centrifuge tubes 50 mL (Labcon, catalog number: 3181-345)
Round-bottom micro tube 1.7 mL (BIO-BIK, catalog number: RC-0170)
Siliconized microcentrifuge tube 1.5 mL (WATSON, catalog number: 131-615CH)
FastGeneTM 0.2 mL 8 strips PCR tube and FlatCap (NIPPON Genetics, catalog number: FG-028FC)
Long tip low adsorption 10 μL (BMBio ST, catalog number: BMSSCH0001)
FastGeneTM tip 200 μL (NIPPON Genetics, catalog number: FG-3001)
Long tip low adsorption 1,000 μL (BMBio ST, catalog number: BMSSCH0004)
Siliconized tip 1,000 μL (WATSON, catalog number: 121-814CH)
Immobilon®-P PVDF membrane (Millipore, catalog number: IPVH00010)
Chromatography paper (ATTO, catalog number: CB-20A)
Equipment
PIPETMAN P-10 (Gilson, catalog number: F144802)
PIPETMAN P-20 (Gilson, catalog number: F123600)
PIPETMAN P-200 (Gilson, catalog number: F123601)
PIPETMAN P-1000 (Gilson, catalog number: F123602)
Pipette Mate NEO (NICHIRYO, catalog number: 00-PMNEO)
Bioclean Bench (PHCbi, catalog number: MCV-B161S)
CO2 incubator Prescyto MG-71C (TAITEC, catalog number: 0081460-000)
Inverted phase contrast microscope (OLYMPUS, catalog number: CK40)
Thermo Minder SDminiN (TITEC, catalog number: 0068750-000)
Aspirator (NISSIN, catalog number: NVP-11)
High-speed refrigerated micro centrifuge (TOMY, catalog number: MX-305)
Biomedical cooler, 4 °C (NIHON FREEZER, catalog number: UKS-5410DHC)
Biomedical freezer, -20 °C (PHCbi, catalog number: MDF-U539)
Ultra-low freezer, -80 °C (PHCbi, catalog number: MDF-DU702VHS1)
VORTEX-GENIE 2 Mixer (M&S Instruments, catalog number: SI-0286)
Ice On I (SK BIO International, catalog number: IO-1)
DynaMagTM-Spin Magnet (Invitrogen, catalog number: 12320D)
Miniature rocker shaker (NISSIN, catalog number: NX-12)
Rotary Mixer with Oshiri Penpen A type (NISSIN, catalog number: NRC-20D)
Hybridization rotator (NISSIN, catalog number: SN-06BN)
Dry thermos unit DTU-1CN (TITEC, catalog number: 0075930-000)
MiniSlab, AE-6401 Gel caster included (ATTO, catalog number: WSE-1165W)
HorizeBLOT 2M (ATTO, catalog number: WSE-4025M)
ImageQuant LAS 4000 mini (Cytiva)
NanoDrop 1000 (Thermo Fischer Scientific, catalog number: ND-1000)
Microchip electrophoresis system for DNA/RNA analysis MultiNA (SHIMADZU, catalog number: MCE-202)
Agencourt SPRIPlate 96R super magnet plate (Beckman Coulter, catalog number: A32782)
SpectraMax Paradigm multi-mode microplate reader (Molecular Devices)
Software and datasets
SLAM-DUNK v.0.4.3 (https://github.com/t-neumann/slamdunk) [12]
R v.4.2.1 (https://www.r-project.org/)
Python v.3.7 (https://www.python.org/)
DESeq2 v.1.3.8 (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html) [13]
deltaTE (https://github.com/SGDDNB/translational_regulation) [14]
ImageQuantTM TL v.1.2 (Cytiva)
Procedure
文章信息
稿件历史记录
提交日期: Jun 27, 2024
接收日期: Aug 30, 2024
在线发布日期: Sep 25, 2024
出版日期: Oct 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Imai, H. and Yamashita, A. (2024). Metabolic RNA Labeling and Translating Ribosome Affinity Purification for Measurement of Nascent RNA Translation. Bio-protocol 14(20): e5091. DOI: 10.21769/BioProtoc.5091.
分类
分子生物学 > RNA > RNA 标记
分子生物学 > RNA > mRNA 转译
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













