- EN - English
- CN - 中文
Detecting Native Protein–Protein Interactions by APEX2 Proximity Labeling in Drosophila Tissues
利用APEX2邻位标记技术检测果蝇组织中的天然蛋白质相互作用
发布: 2024年10月20日第14卷第20期 DOI: 10.21769/BioProtoc.5090 浏览次数: 2149
评审: Hong-Wen TangAnonymous reviewer(s)
Abstract
Enzyme-catalyzed proximity labeling is a potent technique for the discernment of subtle molecular interactions and subcellular localization, furnishing contextual insights into the protein of interest within cells. Although ascorbate peroxidase2 (APEX2) has proven effective in this approach when overexpressed, its compatibility with endogenous proteins remains untested. We improved this technique for studying native protein–protein interactions in live Drosophila ovary tissue. Through CRISPR/Cas9 genome editing, APEX2 was fused with the endogenous dysfusion gene. By pre-treating the tissue with Triton X-100 to enhance biotin-phenol penetration, we successfully identified multiple proteins interacting with the native Dysfusion proteins that reside on the inner nuclear membrane. Our protocol offers a comprehensive workflow for delineating the interactome networks of ovarian components in Drosophila, aiding future studies on endogenous protein–protein interactions in various tissues of other animals.
Key features
• Elucidating Protein-protein interactions provides deeper insights into the regulation of gene expression in molecular network and complex signaling pathways, advancing protein engineering and drug design
• This protocol utilizes CRISPR/Cas9 knock-in technology to tag endogenous proteins with the APEX2 to label nearby proteins within a 20 nm radius in Drosophila melanogaster
• We optimize APEX2-proximity labeling by using Triton X-100 pre-treatment to enhance biotin-phenol penetration into Drosophila ovaries, enabling endogenous proteins enrichment under native conditions
Keywords: Proximity labeling (邻位标记)Graphical overview
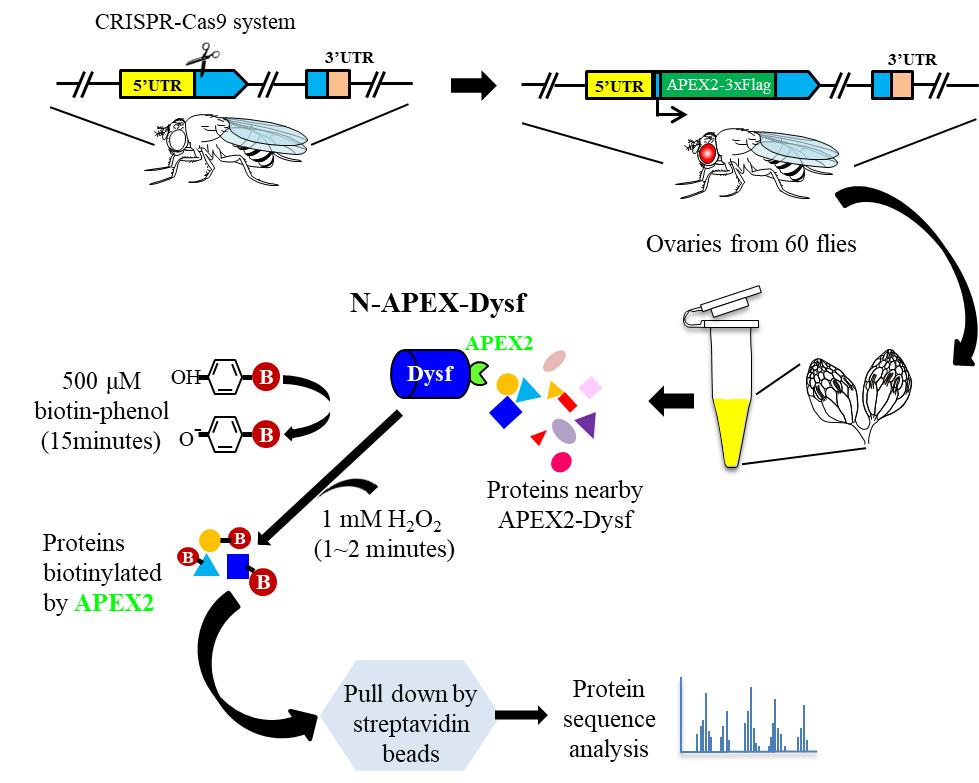
Workflow of proximity labeling with APEX2 tagging endogenous proteins in Drosophila ovary
Background
Proximity labeling is a powerful technique applied to mark adjacent proteins, providing valuable insights into the spatial arrangement of a given protein of interest within cells [1]. This approach excels over conventional assays because it can detect subtle or temporary protein–protein interactions (PPI) and pinpoint the precise localization within intact cellular compartments. Ascorbate peroxidase2 (APEX2) is engineered to enhance proximity labeling techniques for uncovering intricate subcellular proteomes in vivo [2]. By utilizing hydrogen peroxide (H2O2), APEX2 catalyzes biotin-phenol (BP) into biotin-phenoxyl radical, a highly reactive and short-lived species (lasting less than 1 ms), which quickly forms covalent bonds with nearby proteins within a 20 nm radius [3]. The resulting biotinylated proteins can be purified with streptavidin beads and then subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) for identification [3]. Although APEX2 has shown efficacy in cell culture systems and Drosophila tissues when overexpressed, the compatibility with endogenous proteins tagged with this enzyme has not been tested [2–6]. While ectopic expression facilitates research on PPI, it may hinder embryonic development or lead to misinterpretation due to aberrant cellular behavior; a preferable method for unveiling PPI is to purify proteins expressed under native conditions.
In this protocol, we improved APEX2-based proximity labeling in live Drosophila ovary tissue to study PPI in the natural state. By using CRISPR/Cas9 genome editing, we generated a transgenic fly line that carries the APEX2 gene inserted at the N-terminal of endogenous dysfusion (dysf) locus. To enhance protein biotinylation catalyzed by APEX2, fly ovary tissues were pre-treated with Triton X-100 to facilitate the penetration of BP into tissues. After a sequence of protein enrichment and LC-MS/MS analysis, we identified numerous proteins that were labeled by APEX2-tagged Dysf proteins [7].
Altogether, our protocol provides a proximity-labeling workflow for mapping the interactome networks of Drosophila ovary. The level of detail in this protocol shall empower future researchers to explore endogenous PPI in living tissues of other multicellular organisms.
Materials and reagents
Biological materials
Drosophila melanogaster
Schneider's Drosophila medium (Thermo Fisher Scientific, catalog number: 21720024)
Fetal bovine serum (FBS) (Thermo Fisher Scientific, catalog number: A5670701)
100× Penicillin-Streptomycin (Thermo Fisher Scientific, catalog number: 15070063)
Streptavidin-horseradish peroxidase (streptavidin-HRP) (Life Technologies, catalog number: S-911)
cOmpleteTM, EDTA-free protease inhibitor cocktail (Merck, catalog number: 4693132001)
(3aS,4S,6aR)-hexahydro-N-[2-(4-hydroxyphenyl)ethyl]-2-oxo-1H-thieno[3,4-d]imidazole-4-pentanamide (Biotin-phenol, BP) (Iris Biotech, catalog number: LS-3500)
PierceTM Streptavidin magnetic beads (Thermo Fisher Scientific, catalog number: 88816)
Trolox (Merck, catalog number: Al-238813)
Sodium azide (Merck, catalog number: S2002)
Sodium L-ascorbate (Merck, catalog number: A4034)
Hydrogen peroxide solution (H2O2) (Merck, catalog number: 31642)
Phosphate buffered saline (PBS) (Merck, catalog number: P3813)
Bio-Rad protein assay dye reagent concentrate (Bio-Rad, catalog number: 5000006)
Tris base (cyrusbioscience, catalog number: 101-77-86-1)
Glycine (cyrusbioscience, catalog number: 101-56-40-6)
NaCl (cyrusbioscience, catalog number: 101-7647-14-5)
SDS (cyrusbioscience, catalog number: 101-151-21-3)
Triton X-100 (JT Baker®, X198-07)
Bovine serum albumin (BSA) (Bioman, catalog number: ALB001)
Dimethyl sulfoxide (DMSO) (Merck, catalog number: D2650)
10× PBS stock
1 M Tris-HCL pH 8.0
5 M NaCl
10% SDS
Dissection medium (see Recipes)
500 mM Biotin-phenol (see Recipes)
100 mM H2O2 (see Recipes)
1 M Sodium ascorbate (see Recipes)
500 mM Trolox (see Recipes)
1 M Sodium azide (see Recipes)
Quencher solution (see Recipes)
25× Protease inhibitor cocktail (see Recipes)
20% Triton X-100 (see Recipes)
RIPA lysis buffer (see Recipes)
0.3% PBST (see Recipes)
Dissection medium
Reagent Final concentration Quantity or Volume Schneider's Drosophila medium 500 mM 445 mL FBS 10% 50 mL 100× Penicillin-Streptomycin 1× 5 mL Total n/a 500 mL 500 mM Biotin-phenol
Reagent Final concentration Quantity or Volume Biotin-phenol 500 mM 100 mg DMSO n/a Up to 550 μL Total n/a 550 μL The stock needs to be shaken by vortexing.
Aliquot the stock solution into small volumes and store at -80 °C to prevent repeated freeze-thaw cycles.
100 mM H2O2
Reagent Final concentration Quantity or Volume 30% H2O2(10M) 100 mM 10 μL 10× PBS 1× 100 μL H2O n/a 890 μL Total n/a 1 mL Do not store this stock.
1 M Sodium ascorbate
Reagent Final concentration Quantity or Volume Sodium ascorbate 1 M 0.59 g H2O n/a Up to 5 mL Total (optional) n/a 5 mL 500 mM Trolox
Reagent Final concentration Quantity or Volume Trolox 500 mM 12.5145 mg DMSO n/a Up to 100 μL Total (optional) n/a 100 μL Do not store this stock.
1 M Sodium azide
Reagent Final concentration Quantity or Volume Sodium azide 1 M 0.65 g H2O n/a Up to 10 mL Total (optional) n/a 10 mL Aliquots can be stored at -20 °C or below for several months.
Quencher solution
Reagent Final concentration Quantity or Volume 500 mM Trolox 5 mM 10 μL 1 M Sodium azide 10 mM 10 μL 1 M Sodium ascorbate 10 mM 10 μL 10× PBS
H2O
1×
n/a
100 μL
870 μL
Total (optional) n/a 1 mL Make this solution immediately before it is to be used to quench the biotinylation reaction.
Do not store this solution.
25× Protease inhibitor cocktail
Reagent Final concentration Quantity or Volume cOmpleteTM, EDTA-free protease inhibitor cocktail 25× 1 tablet H2O n/a Up to 2 mL Total (optional) n/a 2 mL 20% Triton X-100
Reagent Final concentration Quantity or Volume Triton X-100 20% 10 mL H2O n/a 40 mL Total (optional) n/a 50 mL RIPA lysis buffer
Reagent Final concentration Quantity or Volume 1 M Tris-HCL pH 8.0 50 mM 2.5 mL 5 M NaCl 150 mM 1.5 mL 10% SDS 0.1% 0.5 mL 20% Triton X-100 1% 2.5 mL 25× Protease inhibitor cocktail 1× 2 mL H2O n/a 41 mL Total (optional) n/a 50 mL 0.3% PBST
Reagent Final concentration Quantity or Volume 20% Triton X-100 0.3% 0.75 mL 10× PBS 1× 5 mL H2O n/a 44.25 mL Total (optional) n/a 50 mL
Equipment
Standard equipment for fly incubation
Micro refrigerated centrifuge (Kubota, model: 3740)
Magnetic stand (G-Biosciences, catalog number: 786-888)
Multi-mixer overhead mixer shaker Mischer Rotator (NanoEnTek, model: SLRM-3)
Ultrasonic processor (ChromTech, model: UP-500)
xMarkTM microplate absorbance spectrophotometer (BIO-RAD, catalog number: 1681150)
Mini Trans-Blot electrophoretic transfer cell (BIO-RAD, catalog number: 1703930)
Mini-PROTEAN® Tetra Cell (BIO-RAD, catalog number: 1658001)
Software and datasets
NCBIprot (20180429)
Procedure
文章信息
稿件历史记录
提交日期: May 13, 2024
接收日期: Aug 25, 2024
在线发布日期: Sep 25, 2024
出版日期: Oct 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Wu, J., Wang, C., Hong, W. Y., Jang, A. C. C. and Chang, Y. (2024). Detecting Native Protein–Protein Interactions by APEX2 Proximity Labeling in Drosophila Tissues. Bio-protocol 14(20): e5090. DOI: 10.21769/BioProtoc.5090.
- Wu, J. W., Wang, C. W., Chen, R. Y., Hung, L. Y., Tsai, Y. C., Chan, Y. T., Chang, Y. C. and Jang, A. C. (2022). Spatiotemporal gating of Stat nuclear influx by Drosophila Npas4 in collective cell migration. Sci Adv. 8(29): eabm2411. https://doi.org/10.1126/sciadv.abm2411
分类
发育生物学 > 形态建成 > 能动性
分子生物学 > 蛋白质 > 蛋白质-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












