- EN - English
- CN - 中文
Improving Stability of Spiroplasma citri MreB5 Through Purification Optimization and Structural Insights
通过纯化优化和结构解析提高柑桔螺原体MreB5的稳定性
发布: 2024年10月20日第14卷第20期 DOI: 10.21769/BioProtoc.5086 浏览次数: 1756
评审: Joana Alexandra Costa ReisAnonymous reviewer(s)

相关实验方案

适用于 LC–MS/MS 蛋白质组学分析的大体积细胞培养上清分泌组样品制备方法优化
Basil Baby Mattamana [...] Peter Allen Faull
2025年12月20日 1151 阅读
Abstract
MreB is a prokaryotic actin homolog. It is essential for cell shape in the majority of rod-shaped cell-walled bacteria. Structural and functional characterization of MreB protein is important to understand the mechanism of ATP-dependent filament dynamics and membrane interaction. In vitro studies on MreBs have been limited due to the difficulty in purifying the homogenous monomeric protein. We have purified MreB from the cell-wall-less bacteria Spiroplasma citri, ScMreB5, using heterologous expression in Escherichia coli. This protocol provides a detailed description of purification condition optimization that led us to obtain high concentrations of stable ScMreB5. Additionally, we have provided a protocol for detecting the presence of monovalent ions in the ScMreB5 AMP-PNP-bound crystal structure. This protocol can be used to obtain a high yield of ScMreB5 for carrying out biochemical and reconstitution studies. The strategies used for ScMreB5 show how optimizing buffer components can enhance the yield and stability of purified protein.
Key features
• The protocol is a useful approach to standardize purification of nucleotide-dependent cytoskeletal filaments and other nucleotide-binding proteins.
• The mechanistic basis of how different ions could stabilize a protein, and hence improve yield in purification, has been demonstrated.
• The change in buffer conditions/salt enabled us to get sufficient yield for biochemical and structural characterization.
Keywords: MreB (MreB)Graphical overview
Background
There is an array of prokaryotic filamentous proteins that play a pivotal role in maintaining overall cellular functioning [1]. In the majority of bacteria, complex cell wall synthesis machinery plays a major role in cell shape maintenance. Studies have extensively analyzed how shape is regulated in E. coli and B. subtilis. Genetic studies show that deleting mreB causes loss of rod shape and eventual lysis, indicating the pivotal role of the MreB protein in bacterial cell shape and maintenance [2–4].
MreB forms antiparallel double protofilaments in the presence of nucleotide, ATP, or ADP that align parallel to the long axis of the rod-shaped cells [5]. The processive movement of MreB along with the other components of cell wall synthesis machinery drives the insertion of peptidoglycan, thus enforcing the cell shape [6,7].
MreB5 is responsible for determining helicity and motility in the cell-wall-less bacteria Spiroplasma citri [8]. In our recent work, we reported the structure and biochemical characterization of ScMreB5 and its mutants [9]. We showed that the nucleotide state of ScMreB5 determines the filament dynamics and membrane interaction. MreBs are notoriously challenging to purify because they tend to aggregate when overexpressed. Obtaining monomeric and homogeneous proteins for biochemical and structural studies has been challenging, resulting in few MreBs being characterized in vitro.
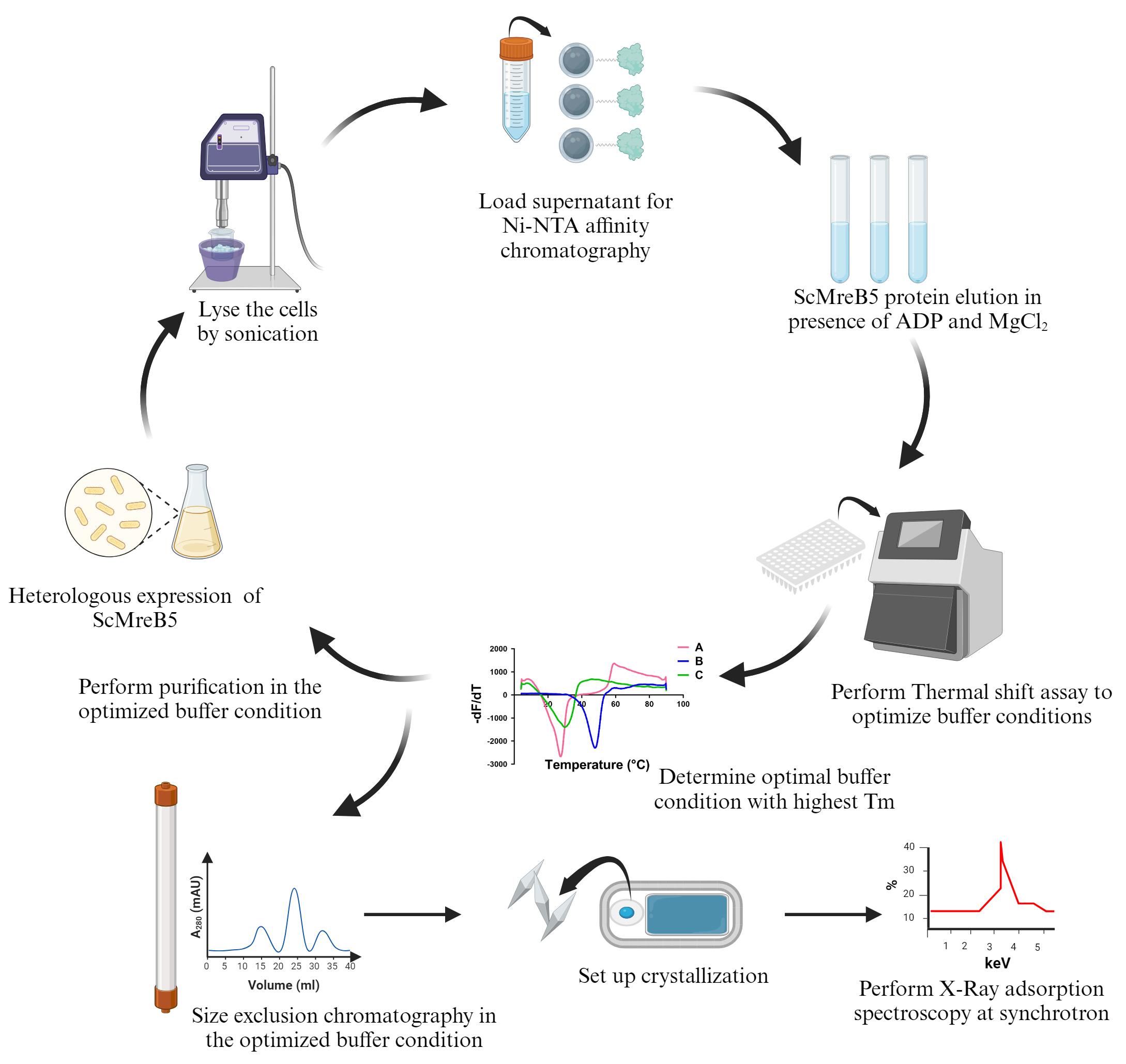
For in vitro characterization and structure determination, the scmreb5 gene was cloned into the pHis17 expression vector, under the T7 promoter, between NdeI and BamHI restriction endonuclease site. The cloned gene was codon-optimized by mutating the TGA codon to TGG, since in S. citri, UGA is recognized as tryptophan [10]. A restriction-free cloning strategy [11] was chosen for cloning, and the clone was confirmed by DNA sequencing. The cloned plasmid was transformed into the BL21-AI expression strain, which has a T7 RNA polymerase tightly regulated by the araBAD promoter and induced with arabinose for overexpression.
This protocol paper describes optimizing ScMreB5 protein purification, where precipitation in NaCl-containing buffer was prevented by adding excess ADP and MgCl2 to stabilize the protein. This addition prevented any further biochemical characterization of ScMreB5. To eliminate this issue, we optimized the buffer conditions for protein purification using a thermal shift assay [12,13]. The optimized buffer containing KCl removed the necessity for ADP and MgCl2 addition during the purification process. To understand the structural significance of KCl in stabilizing the protein, we performed X-ray adsorption spectroscopy on ScMreB5-AMP-PNP crystals. This technique is widely used for detecting metal ions within a protein crystal. Since the X-ray absorption edges of different elements occur at specific energies, it allows the identification of specific metal ions within the crystal. The presence of potassium ion bridging the protein with the nucleotide provided a structural basis for why the stability of ScMreB5 improved in the presence of KCl.
Materials and reagents
For transformation and large culture expression
100 mm Petri dish (Himedia, catalog number: PW1305-1x100NO)
Luria Bertani agar, LB agar (Himedia, catalog number: M1151-1KG)
Luria Bertani broth powder (Himedia, catalog number: M1245-1KG)
BL21-AI chemically competent E. coli cells (Invitrogen, catalog number: C6070-03)
ScMreB5 cloned pHis17 plasmid (Hexa His tag at the C-terminus)
Ampicillin (Sigma-Aldrich, catalog number: A9518-25G)
100 mL conical flask (Borosil)
2 L conical flask (Borosil)
0.22 µm syringe filter (Cytiva, catalog number: WHA-9914-2502)
10 mL syringe for single use (Dispovan, catalog number: JK-10ml-0110)
L-Arabinose (Himedia, catalog number: GRM037-25G)
For purification
150 mL beaker (Borosil, catalog number: 1000D18)
0.5 mL centrifuge tubes (Tarsons, catalog number: 500000)
1.5 mL centrifuge tubes (Tarsons, catalog number: 500010)
0.2 mL PCR tubes (Tarsons, catalog number: 510051)
50 mL conical centrifuge tubes (Tarsons, catalog number: TAR-500041)
100 mL conical flasks (Borosil, catalog number: 5020016)
500 mL conical flasks (Borosil, catalog number: 5020024)
2 L conical flasks (Borosil, Catalog number: 5020030)
Test tubes, 15 × 125 mm (Borosil, catalog number: 9800U04)
HiIndicator pH paper, pH range 6.5–9.0 (Himedia, catalog number: LA321-1PK)
5 mL His-Trap column (Cytiva, catalog number: 17524801)
Bradford 1× dye reagent (Bio-Rad, catalog number: 5000205)
Bovine serum albumin (BSA) (Himedia, catalog number: TC548-5G)
Durapore membrane filter, 0.22 µm, hydrophilic PVDF, 47 mm membrane (Sigma-Aldrich, catalog number: GVWP04700)
Superdex 200 Increase 10/300 GL (Cytiva, catalog number: 28990944)
Oak Ridge centrifuge tubes, 50 mL (Thermo Scientific, catalog number: 3118-0050)
Thick-walled Polycarbonate tubes, 11 × 34 mm, 1 mL (Beckman Coulter, catalog number: 343778)
Hydrochloric acid (Qualigens, catalog number: Q29145)
Tris, free base, for molecular biology (Himedia, catalog number: MB029-5KG)
Glycine (Sigma-Aldrich, catalog number: G8898-1KG)
Sodium dodecyl sulphate (Sigma-Aldrich, catalog number: 436143)
TEMED (Tetramethylethylenediamine) (Sigma-Aldrich, catalog number: T9281-25ML)
Ammonium persulphate (Sigma-Aldrich, catalog number: A3678)
Acrylamide (Sigma-Aldrich, catalog number: A8887-500G)
Bis-acrylamide (Sigma-Aldrich, catalog number: 146072-500G)
0.75 mm spacer plate (Bio-Rad, catalog number: 1653310)
Short plates (Bio-Rad, catalog number: 1653308)
10-well comb, 0.75 mm (Bio-Rad, catalog number: 1653354)
15-well comb, 0.75 mm (Bio-Rad, catalog number: 1653355)
Coomassie Brilliant Blue (R-250) (Sigma-Aldrich, catalog number: 27816)
2× Laemmli sample buffer (Bio-Rad, catalog number: 1610737)
DTT (Sigma-Aldrich, catalog number: D9779-25G)
Ethanol for molecular biology (Omnis)
Glacial acetic acid (Qualigens, catalog number: Q11007)
Sodium chloride (NaCl) (Qualigens, catalog number: Q15918)
Potassium chloride (KCl) (Qualigens, catalog number: Q13305)
Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266-100G)
ADP adenosine 5'-diphosphate sodium salt (Sigma-Aldrich, catalog number: A2754-500MG)
Imidazole (Sigma-Aldrich, catalog number: I2399-500G)
Glycerol (Fisher Scientific, catalog number: 15457)
Dialysis tubing 10 kDa MWCO (Thermo Fisher Scientific, catalog number: 68100)
Dialysis tubing clamps (Sigma-Aldrich, catalog number: Z371092-10EA)
Vivaspin® Turbo 15 PES 10 kDa MWCO (Sartorius, catalog number: VS15T01)
Precision Plus Protein Dual Color Standards (Bio-Rad, catalog number: 1610394)
Micro test plate, flat Bottom (Tarsons, catalog number: 941196)
Liquid nitrogen
For thermal shift assay
MultiplateTM 96-well PCR plates, low profile, unskirted, white (Bio-Rad, catalog number: MLL9651)
Microseal 'B' PCR plate sealing film, adhesive, optical (Bio-Rad, catalog number: MSB1001)
SYPRO orange (Sigma-Aldrich, catalog number: S5692)
For crystallization
48-well crystallization plate (Hampton Research, catalog number: HR3-180)
Sodium phosphate monobasic monohydrate (Sigma-Aldrich, catalog number: 71507-250G)
Sodium phosphate dibasic (Sigma-Aldrich, catalog number: S9763-100G)
AMP-PNP Adenosine 5'-(β, γ-imido) triphosphate lithium salt hydrate (Sigma-Aldrich, catalog number: 10102547001)
Magnesium chloride (details mentioned above)
PEG 3350 (Sigma-Aldrich, catalog number: 202444-500G)
Glycerol (Sigma-Aldrich, catalog number: G5516)
CrystalCap SPINE HT, 0.05-0.1 mm CryoLoop (Hampton Research, catalog number: HR8-120)
Stock solutions (see Recipes)
100 mg/mL ampicillin
1 mg/mL BSA
2 M MgCl2
1 M unbuffered Tris
100 mM ADP
100 mM AMP-PNP
20% Arabinose (w/v)
5 M NaCl
3 M KCl
2 M Tris pH 8
2 M Imidazole
30% Acrylamide solution (w/v)
10% Ammonium persulphate (w/v)
10% SDS
1.5 M Tris pH 6.8
1.5 M Tris, pH 8.8
1 M sodium phosphate monobasic monohydrate
1 M sodium phosphate dibasic
1 M sodium phosphate buffer, pH 7.8
40% PEG 3350 (w/v)
20% Glycerol (v/v)
50× SYPRO Orange
Solutions and buffers for purification (see Recipes)
10× TGS buffer for SDS-PAGE gels
SDS-PAGE staining solution
SDS-PAGE destaining solution
5% SDS-PAGE stacking gel
12% SDS-PAGE resolving gel
Lysis buffer (Na)
Buffer A (Na)
Buffer B (Na)
Storage buffer
Lysis buffer (K)
Buffer A (K)
Buffer B (K)
Crystallization conditions
Recipes
Materials to be autoclaved
LB Agar
Dissolve 4 g of LB Agar in RO (reverse osmosis) water to a final volume of 100 mL in a 500 mL conical flask. Autoclave at 121 °C for 15 min at 2.8 bar pressure for sterilization. Once cooled enough to touch, add 100 µL of 100 mg/mL ampicillin solution to the LB agar and gently stir. Pour 25 mL of LB agar into the Petri plates. Store the plates at 4 °C after the LB agar solidifies.
1× LB Broth, 30 mL
Dissolve 0.75 g of Luria Bertani broth powder in RO water to a final volume of 30 mL in a 100 mL conical flask. Autoclave at 121 °C for 15 min at 2.8 bar pressure for sterilization.
1× LB Broth, 1,000 mL
Dissolve 25 g of Luria Bertani broth powder in RO water to a final volume of 1,000 mL in a 2 L conical flask. Autoclave at 121 °C for 15 min at 2.8 bar pressure for sterilization.
15 × 12.5 mm test tubes
Autoclave 20–25 clean test tubes at 121 °C for 15 min at 2.8 bar pressure for sterilization.
Stock solutions
100 mg/mL Ampicillin
Dissolve 0.5 g of ampicillin powder in ultrapure water to a final volume of 5 mL. Sterilize the solution using a 0.22 µm syringe filter.
1 mg/mL BSA
Dissolve 1 mg of BSA powder in ultrapure water to a final volume of 1 mL. Use this solution to prepare standards (0.1–1.0 mg/mL) in ultrapure water for protein concentration estimation using Bradford reagent.
2 M MgCl2
Dissolve 1.90 g of MgCl2 powder in ultrapure water to a final volume of 10 mL. Filter the solution using a 0.22 µm syringe filter.
1 M unbuffered Tris
Dissolve 1.21 g of unbuffered Tris-base powder in ultrapure water to a final volume of 10 mL. Filter the solution using a 0.22 µm syringe filter.
100 mM ADP
Dissolve 23.5 mg of ADP powder in 100 µL of ultrapure water. Adjust the pH to 7–7.5 with 1 M unbuffered Tris using a pH strip. Make up the volume to 250 µL.
100 mM AMP-PNP
Dissolve 5 mg of ADP powder in 30 µL of ultrapure water. Adjust the pH to 7–7.5 with 1 M unbuffered Tris using a pH strip. Make up the volume to 100 µL.
20% (w/v) Arabinose
Dissolve 10 g of arabinose in RO water to a final volume of 50 mL. Filter the solution using a 0.22 µm syringe filter.
5 M NaCl
Dissolve 146.1 g of NaCl powder in RO water to a final volume of 500 mL. Filter the solution using a vacuum filter assembly with a 0.22 µm membrane filter.
3 M KCl
Dissolve 111.8 g of KCl powder in RO water to a final volume of 500 mL. Filter the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane).
2 M Tris, pH 8
Dissolve 60.7 g of Tris-base in 120 mL of RO water. Adjust the pH to 8 using concentrated hydrochloric acid using a pH meter. Make up the volume to 250 mL. Filter the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane).
Note: Follow the required lab safety procedures while handling hydrochloric acid.
2 M Imidazole
Dissolve 68.07 g of imidazole powder in 500 mL of RO water. Filter the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane).
30% Acrylamide solution
Dissolve 146 g of acrylamide and 3.9 g of bis-acrylamide powders in 500 mL of RO water. Filter the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane).
10% Ammonium persulphate
Dissolve 5 g of ammonium persulphate powder in RO water to a final volume of 50 mL. Filter the solution using a 0.22 µm syringe filter.
10% SDS
Dissolve 5 g of SDS powder in RO water to a final volume of 50 mL.
1.5 M Tris pH 6.8
Dissolve 45.5 g of Tris-base in 120 mL of RO water. Adjust the pH to 6.8 using concentrated hydrochloric acid and a pH meter. Make up the volume to 250 mL. Filter the buffer using a 0.22 µm syringe filter.
1.5 M Tris, pH 8.8
Dissolve 45.5 g of Tris-base in 120 mL of RO water. Adjust the pH to 8.8 using concentrated hydrochloric acid and a pH meter. Make up the volume to 250 mL. Filter the buffer using a 0.22 µm syringe filter.
1 M sodium phosphate monobasic monohydrate
Dissolve 1.37 g of sodium phosphate monobasic monohydrate powder in ultrapure water to a final volume of 10 mL. Filter the solution using a 0.22 µm syringe filter.
1 M sodium phosphate dibasic
Dissolve 1.42 g of sodium phosphate dibasic powder in ultrapure water to a final volume of 10 mL. Filter the solution using a 0.22 µm syringe filter.
1 M sodium phosphate buffer, pH 7.8
Titrate sodium phosphate monobasic monohydrate using 1 M sodium phosphate dibasic to obtain pH 7.8. Use HiIndicator pH Paper, pH range 6.5–9.0 for adjusting the pH.
40% PEG 3350
Dissolve 20 g of PEG 3350 powder in ultrapure water to a final volume of 50 mL. Filter the solution using a 0.22 µm syringe filter. Use this stock to make 16% PEG 3350.
20% Glycerol
Mix 2 mL of glycerol with 8 mL of ultrapure water. Filter the solution using a 0.22 µm syringe filter.
50× SYPRO Orange
Mix 1 µL of 5,000× SYPRO Orange in 9 µL of DMSO to make 500× SYPRO orange. Dilute 500× SYPRO orange to 50× by mixing 2 µL of 500× SYPRO orange in 18 µL in ultrapure water.
Buffers and working stock solutions
10× TGS Buffer
Dissolve 30 g of Tris-base, 144 g of glycine, and 10 g of SDS in 1,000 mL of RO water. Use it to make 1× TGS buffer for running the SDS-PAGE gels.
Coomassie staining solution
Dissolve 2.5 g of Coomassie Brilliant Blue (R-250) in 500 mL of ethanol by stirring. Add 100 mL of glacial acetic acid and 400 mL of RO water to make the final volume of 1,000 mL.
De-staining solution
Mix 100 mL of glacial acetic acid and 200 mL of ethanol with 700 mL of RO water.
12% SDS-PAGE resolving gel (5 mL)
Mix the following solutions and pour onto the 0.75 mm clamped gel setup.
RO water (1.6 mL)
30% acrylamide solution (2 mL)
Tris-HCl, pH 8.8 (1.5 mL)
10% SDS (50 µL)
10% ammonium persulphate (50 µL)
TEMED (3 µL)
Add 100 µL of 100% isopropanol to even out the layer of resolving gel. Once solidified, carefully tilt the gel setup to one side and use tissue paper to remove isopropanol.
5% SDS-PAGE stacking gel (1 mL)
Mix the following solutions and pour over the solidified resolving gel.
RO water (680 µL)
30% acrylamide solution (170 µL)
Tris-HCl, pH 6.8 (130 µL)
10% SDS (10 µL)
10% ammonium persulphate (10 µL)
TEMED (1 µL)
After pouring the stacking solution, place a 10- or 15-well 0.75 mm comb.
Lysis buffer (Na)
200 mM NaCl (10 mL from 5 M NaCl)
50 mM Tris-HCl pH 8 (6.25 mL from 2 M Tris-HCl pH 8)
10% glycerol (12.5 mL from 100% glycerol)
Make up the volume to 250 mL with RO water. Filter and degas the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane). Pre-chill the buffer at 4 °C for purification.
Buffer A (Na)
Used as binding buffer in Ni-NTA chromatography.
200 mM NaCl (20 mL from 5 M NaCl)
50 mM Tris-HCl pH 8 (12.5 mL from 2 M Tris-HCl pH 8)
Make up the volume to 500 mL with RO water. Filter and degas the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane). Pre-chill the buffer at 4 °C for purification.
Buffer B (Na)
Used as elution buffer in Ni-NTA chromatography.
200 mM NaCl (20 mL from 5 M NaCl)
50 mM Tris-HCl pH 8 (12.5 mL from 2 M Tris-HCl pH8)
500 mM imidazole (125 mL from 2 M imidazole)
Make up the volume to 500 mL with RO water. Filter and degas the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane). Pre-chill the buffer at 4 °C before using for purification.
Storage buffer
Used for buffer exchange and protein storage.
50 mM NaCl (1 mL from 5 M NaCl)
50 mM Tris-HCl pH 8 (2.5 mL from 2 M Tris-HCl pH 8)
Make up the volume to 100 mL with RO water. Filter and degas the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane). Pre-chill the buffer at 4 °C for purification.
Lysis buffer (K)
300 mM KCl (25 mL from 3 M KCl)
50 mM Tris-HCl pH 8 (6.25 mL from 2 M Tris-HCl pH 8)
10% glycerol (12.5 mL from 100% glycerol)
Make up the volume to 250 mL with RO water. Filter and degas the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane). Pre-chill the buffer at 4 °C for purification.
Buffer A (K)
Used as binding buffer in Ni-NTA chromatography and buffer for size exclusion chromatography.
300 mM KCl (100 mL from 3 M KCl)
50 mM Tris-HCl pH 8 (25 mL from 2 M Tris-HCl pH 8)
Make up the volume to 1000 mL with RO water. Filter and degas the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane). Pre-chill the buffer at 4 °C for purification.
Buffer B (K)
Used as elution buffer in Ni-NTA chromatography.
300 mM KCl (50 mL from 3 M KCl)
50 mM Tris-HCl pH 8 (12.5 mL from 2 M Tris-HCl pH 8)
500 mM imidazole (125 mL from 2 M imidazole)
Make up the volume to 500 mL with RO water. Filter and degas the solution using a vacuum filter assembly with DuraporeTM membrane filter (0.22 µm, hydrophilic PVDF, 47 mm membrane). Pre-chill the buffer at 4 °C before using for purification.
Crystallization condition for X-ray fluorescence scanning
0.15 M sodium phosphate buffer, pH 7.8 (150 µL from 1 M sodium phosphate buffer, pH 7.8)
16% PEG 3350 (400 µL from 40% PEG 3350)
Make up the volume to 1000 µL with ultrapure water. Sterilize using a 0.22 µm syringe filter.
Equipment
Shaker incubator (Thermo Scientific, model: MaxQTM 6000)
Eppendorf® ThermoMixer® F2.0 (EP5387000013)
Sonicator (Sonics VibraCell)
E-GelTM imager system (Fisher Scientific, catalog number: 4466611)
pH meter (Thermo Scientific, model: ORION STAR A111)
Vacuum filter assembly (Millipore, model: XX1014720)
Centrifuge (Avanti J26S-XP Non-IVD)
Tabletop ultracentrifuge (Beckman Coulter Optima MAX-XP)
High-speed centrifuge (Eppendorf, model: 5430 R)
Benchtop centrifuge (Eppendorf, model: 5810 R)
Centrifuge (Eppendorf MiniSpin®)
ÄKTA PrimeTM Plus FPLC system
Bio-Rad NGCTM chromatography system QuestTM 10
SDS-PAGE apparatus (Bio-Rad, catalog number: 1658002FC)
CLARIOstar® microplate reader
Bio-Rad CFX96 Touch Real-Time PCR Detection System
Software and datasets
GraphPad Prism (for analyzing and plotting thermal shift assay and size exclusion chromatography)
Microsoft Excel (for exporting thermal shift assay results)
ImageJ (for SDS-PAGE gel)
Procedure
文章信息
稿件历史记录
提交日期: Apr 18, 2024
接收日期: Aug 23, 2024
在线发布日期: Sep 28, 2024
出版日期: Oct 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Pande, V. and Gayathri, P. (2024). Improving Stability of Spiroplasma citri MreB5 Through Purification Optimization and Structural Insights. Bio-protocol 14(20): e5086. DOI: 10.21769/BioProtoc.5086.
- Pande, V., Mitra, N., Bagde, S. R., Srinivasan, R. and Gayathri, P. (2022). Filament organization of the bacterial actin MreB is dependent on the nucleotide state. J Cell Biol. 221(5): e202106092.
分类
生物化学 > 蛋白质 > 分离和纯化
细胞生物学 > 细胞结构 > 细胞质膜
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link