- EN - English
- CN - 中文
Iterative Immunostaining and NEDD Denoising for Improved Signal-To-Noise Ratio in ExM-LSCM
通过迭代免疫染色与NEDD去噪提升ExM-LSCM的信噪比
(*contributed equally to this work) 发布: 2024年09月20日第14卷第18期 DOI: 10.21769/BioProtoc.5072 浏览次数: 2625
评审: Xiaoyi ZhengLei GaoDevashish DWIVEDI
Abstract
Expansion microscopy (ExM) has significantly reformed the field of super-resolution imaging, emerging as a powerful tool for visualizing complex cellular structures with nanoscale precision. Despite its capabilities, the epitope accessibility, labeling density, and precision of individual molecule detection pose challenges. We recently developed an iterative indirect immunofluorescence (IT-IF) method to improve the epitope labeling density, improving the signal and total intensity. In our protocol, we iteratively apply immunostaining steps before the expansion and exploit signal processing through noise estimation, denoising, and deblurring (NEDD) to aid in quantitative image analyses. Herein, we describe the steps of the iterative staining procedure and provide instructions on how to perform NEDD-based signal processing. Overall, IT-IF in ExM–laser scanning confocal microscopy (LSCM) represents a significant advancement in the field of cellular imaging, offering researchers a versatile tool for unraveling the structural complexity of biological systems at the molecular level with an increased signal-to-noise ratio and fluorescence intensity.
Key features
• Builds upon the method developed by Mäntylä et al. [1] and introduces the IT-IF method and signal-processing platform for several nanoscopy imaging applications.
• Retains signal-to-noise ratio and significantly enhances the fluorescence intensity of ExM-LSCM data.
• Automatic estimation of noise, signal reconstruction, denoising, and deblurring for increased reliability in image quantifications.
• Requires at least seven days to complete.
Keywords: Iterative indirect immunofluorescence staining (迭代间接免疫荧光染色)Graphical overview
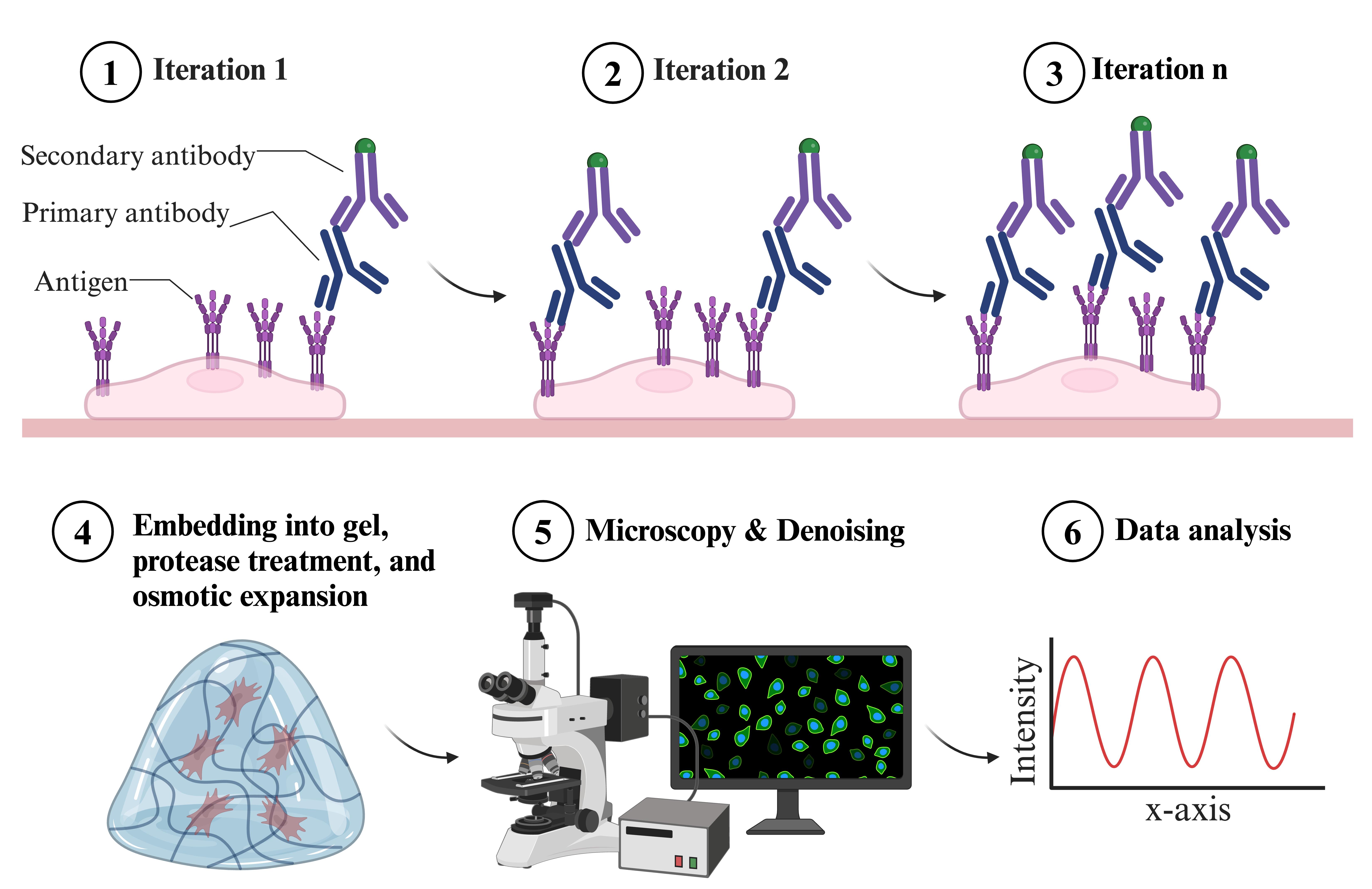
Overview of indirect iterative immunofluorescence staining (IT-IF) procedure
Background
Expansion microscopy (ExM) has emerged as a tempting super-resolution microscopy technique for fluorescence imaging of cell and tissue organization. Along with other super-resolution modalities, ExM now extends optical imaging to the nanoscale, yielding unprecedented biological insights within molecularly crowded environments such as the nucleus. In ExM, fixed and gel-embedded samples are isotropically expanded in water by 4× to 10×, allowing imaging beyond the classical diffraction limit of light microscopy and resolving structures in near-molecular resolution. As a result, ExM allows a lateral resolution of ~70 nm with laser scanning confocal microscope (LSCM); the more recently developed iterative ExM including a second gel step allows 10× improvement, achieving a resolution of ~25 nm [2]. Recently, ExM has also been optimized for clinical specimens. The expansion pathology has been speculated to enable routine nanoscale imaging of pathological samples aiding clinical research [3].
However, ExM imaging of immunofluorescence-based samples has drawbacks related to technical difficulties in obtaining the optimal density of the fluorophores. ExM imaging of the samples results in a substantially weaker signal due to the dilution of the fluorophores caused by the sample expansion; the dilution scales to the third power of the (linear) expansion factor. Thus, expansion significantly reduces image brightness and creates noisy images with low signal-to-noise ratio (SNR). Conventional indirect immunostaining can be insufficient, leading to low-density labeling of the targets and, depending on the antibody type, often includes marked off-target staining of the background, also contributing to low image contrast and signal-to-background ratio (SBR) [4,5]. The accessibility of the target epitopes can be reduced, further lowering the image quality [6,7]. Also, super-resolution imaging methods pushing this barrier, such as structured illumination microscopy, pose challenges to the sample properties. When imaging biological samples in the sub-diffraction scale, low labeling density can lead to long acquisition times, higher excitation laser powers, and higher detector sensitivities, leading to increased noise and photobleaching.
We addressed these problems and developed an indirect iterative immunostaining (IT-IF) method for fluorescent labeling of ExM samples. The IT-IF counteracting the expansion-induced decrease in sample fluorescent intensity and signal quality significantly improves the sufficient staining of molecular structures aiding in structural analyses. We argue that the IT-IF approach facilitates nanoscopy, the detection and quantitative analysis of sub-resolution-sized molecular structures, especially within the nucleus. We show how post-image processing through noise estimation, denoising, and deblurring (NEDD) improves the low signal quality in ExM [8] even better than conventional deconvolution and provide step-by-step instructions on how to use the NEDD image processing tool to perform noise reduction on LSCM data. We prove that IT-IF leads to increased signal intensity without compromising the SBR, advancing super-resolution imaging of highly compact intranuclear structures. Finally, we exploit these methods to reveal nanoscopic structural details of nuclear lamina network organization.
Materials and reagents
Reagents
Paraformaldehyde (PFA), 20% (Electron Microscopy Sciences, catalog number: 15713-S)
Primary antibody used in this study: Mouse anti-Lamin A/C (E-1) (Santa Cruz Biotechnology, catalog number: sc-376248)
Secondary antibody used in this study: AlexaTM-conjugated goat anti-mouse IgG (H+L) cross-adsorbed secondary antibody (Alexa FluorTM 488) (Thermo Fisher Scientific, catalog number: A-11008)
TritonTM X-100 (Sigma-Aldrich, Merck, catalog number: X100-100ML)
Bovine serum albumin (BSA) (PAN Biotech, catalog number: P06-139210)
Sodium acrylate (Sigma-Aldrich, Merck, catalog number: 408220)
Acrylamide (0.40 g/mL, 40%) (Bio-Rad Laboratories, catalog number: 1610140)
N,N'-Methylenebisacrylamide (Sigma-Aldrich, Merck, catalog number: 1015460100)
Sodium chloride (Sigma-Aldrich, Merck, CAS #7647-14-5)
N,N,N',N'-Tetramethylethylenediamine (TEMED, 100%) (Sigma-Aldrich, Merck, catalog number: 411019)
Acryloyl X – SE (10 mg/mL in dry DMSO) (Thermo Fisher Scientific, catalog number: A20770)
4-Hydroxy-TEMPO (Sigma-Aldrich, Merck, catalog number: 176141)
Tris base (Sigma-Aldrich, Merck, catalog number: 10708976001)
Proteinase K (New England Biolabs, catalog number: P8107S)
Ammonium persulphate (APS, powder) (Sigma-Aldrich, Merck, catalog number: A3678)
High-melt agarose (1%, w/v) (Thermo Fisher Scientific, catalog number: J61123.22)
Phosphate buffered saline, with Ca2+/Mg2+ (PBS, 10×) (Gibco, Thermo Fisher Scientific, catalog number: J62036.K2)
EDTA, 10 mM (Promega, catalog number: A2631)
Guanidine HCl (8 M) (Thermo Fisher Scientific, catalog number: 24115)
dH2O
Absolute EtOH
Solutions
1× PBS (see Recipes)
Permeabilization solution A (see Recipes)
Permeabilization solution B (see Recipes)
Antibody diluent containing blocking agent (see Recipes)
Tris-HCl, 50 mM, pH 8 (see Recipes)
Sodium chloride, 5 M (see Recipes)
APS, 10% (w/v) (see Recipes)
Monomer solution (see Recipes)
Gelling solution (see Recipes)
Anchoring solution (see Recipes)
Digestion solution (see Recipes)
Recipes
1× PBS
Reagent Final concentration Quantity or Volume 10× PBS 1/10 10 mL dH2O n/a 90 mL Total n/a 100 mL Permeabilization buffer A
Reagent Final concentration Quantity or Volume 1× PBS n/a 50 mL BSA*
Triton X-100
0.5% (w/v)
0.5% (v/v)
0.25 g
250 µL
Total n/a 50 mL *Prepare fresh. The prepared solution can be stored at 4 °C and used for two weeks.
Permeabilization buffer B*
Reagent Final concentration Quantity or Volume 1× PBS n/a 50 mL BSA** 0.5% (w/v) 0.25 g Triton X-100 0.2% (v/v) 100 µL Total n/a 50 mL *Permeabilization buffer B contains a reduced concentration of Triton X-100 for a gentler permeabilization during iteration steps 2–4.
**Prepare fresh. The prepared solution can be stored at 4 °C and used for two weeks.
Antibody diluent containing blocking agent
Reagent Final concentration Quantity or Volume 1× PBS n/a 50 mL BSA 3% (w/v) 1.5 g Total n/a 50 mL Tris-HCl, 50 mM, pH 8
Reagent Final concentration Quantity or Volume Tris base 50 mM 0.606 g dH2O n/a 60 mL Adjust to pH 8.0 with HCl dH2O to 100 mL Total n/a 100 mL Sodium chloride, 5 M
Reagent Final concentration Quantity or Volume Sodium chloride 5 M (0.292 g/mL) 2.922 g dH2O to 10 mL Total n/a 10 mL APS, 10% (w/v)
Reagent Final concentration Quantity or Volume APS powder* 10% (w/v) 1 g dH2O to 10 mL Total n/a 10 mL *Store at room temperature (RT) in a desiccator.
Monomer solution*
Reagent (stock concentration) Final concentration Volume Sodium acrylate (0.38 g/mL)** 0.086 g/mL 2.25 mL Acrylamide (0.40 g/mL, 40%) 0.025 g/mL (2.5%) 0.58 mL N,N'-Methylenebisacrylamide (0.02 g/mL, 2%) 0.0015 g/mL (0.15%) 0.75 mL Sodium chloride (0.292 g/mL, Recipe 6) 0.117 g/mL 4 mL 10× PBS 1× 1 mL dH2O n/a 0.82 mL Total n/a 9.4 mL*** *Can be stored at -20 °C for up to 2 months.
**Sodium acrylate may have a variable purity affecting performance. Ensure it appears colorless under room light. If the reagent appears yellow, discard the stock without using it. Store at -20 °C in a desiccator.
***9.4/10 mL (1.06×), the remaining 6% of the volume will be brought up by the initiator, accelerator, and inhibitor as needed (see below).
Gelling solution*
Reagent Volume Monomer solution** 188 µL dH2O (or inhibitor 4-Hydroxy-TEMPO)*** 4 µL TEMED**** 4 µL APS (10%, w/v) (Recipe 7) 4 µL Total 200 µL *Prepare fresh and pipette reagents in the given order on ice.
**Degas the monomer solution after preparation by using a degassing chamber to remove any air bubbles. Air will impair the homogenous polymerization of the gel.
***OPTIONAL replacement of water. 4-Hydroxy-TEMPO solution appears yellow. It slows down the gelling, giving a wider temporal working window.
**** N,N,N',N'-Tetramethylethylenediamine. Store at RT in a desiccator.
Anchoring solution
Reagent (stock concentration) Volume Acryloyl X – SE (10 mg/mL) 20 µL PBS (1×) 1980 µL Total 2,000 µL (2 mL) Prepare fresh.
Digestion solution
Reagent (stock concentration) Final concentration Volume Tris-HCl pH 8.0 (50 mM, Recipe 5) 50 mM 1,099 µL EDTA (10 mM) 1 mM 140 µL Triton X-100 0.5% (v/v) 7 µL Guanidine HCl (8 M) 0.8 M 140 µL Add before use: Proteinase K 1:100 (8 units/mL) 14 µL Total n/a 1.4 mL
Laboratory supplies
High-performance cover glasses 22 × 22 mm (D = 0.17 mm) (Carl Zeiss, catalog number: 474030-9020-000)
High-performance cover glasses 18 × 18 mm (D = 0.17 mm) (Carl Zeiss, catalog number: 474030-9000-000)
Scalpels
Spoons or spoon spatulas (stainless steel)
Scissors
Tweezers
Pasteur pipettes
Caliper
Hex nut, 9 mm in diameter
Circular glass coverslip, 1.5 mm thickness, 13 mm in diameter (Marienfeld, VWR, catalog number: MARI0117530)
Cell culture Petri dishes, non-treated, 35 mm in diameter
Cell culture Petri dishes, non-treated, 60 mm in diameter
Cell culture 6-well plates
Parafilm cut to 18 mm × 18 mm pieces to coat cover glasses of similar size
Cyanoacrylate super glue for gluing parafilm to cover glasses
In-house 3D-printed spacers with 350 µm thickness [acrylonitrile butadiene styrene (ABS) plastic], see specification of design for 3D printing from Mäntylä et al. [1]
In-house 3D-printed molds for cutting the gels [polylactic acid (PLA) plastic], see specification of design for 3D printing from Mäntylä et al. [1]
Coverslip cell chamber (Aireka Cells, catalog number: SC15022)
1.5 mL Eppendorf tubes
2.0 mL Eppendorf tubes
P1000 pipette tips
P200 pipette tips
P10 pipette tips
Polystyrene box (size L30 cm × H30 cm × W30 cm)
Tinfoil
Ice
Equipment
Vacuum degassing chamber (e.g., Applied Vacuum Engineering, model: DP3, 3 L)
Vacuum pump
Microwave
Inverted laser scanning confocal fluorescence microscope (e.g., Nikon Instruments, model: Nikon A1R in Ti2 Eclipse)
Software and datasets
Fiji/ImageJ image processing package (open source), Fiji (ImageJ, https://imagej.net/software/fiji/) can be used to analyze acquired microscopy images [9])
Noise estimation, denoising, and deblurring (NEDD) software (open source), MATLAB (v1.0.1, release date Apr 17, 2023). The code has been deposited to GitHub: https://github.com/lucioazzari/NoiseEstimationDenoisingDeblurring
Procedure
文章信息
稿件历史记录
提交日期: Jun 14, 2024
接收日期: Aug 8, 2024
在线发布日期: Aug 30, 2024
出版日期: Sep 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Azzari, L., Vippola, M., Nymark, S., Ihalainen, T. O. and Mäntylä, E. (2024). Iterative Immunostaining and NEDD Denoising for Improved Signal-To-Noise Ratio in ExM-LSCM. Bio-protocol 14(18): e5072. DOI: 10.21769/BioProtoc.5072.
- Mäntylä, E., Montonen, T., Azzari, L., Mattola, S., Hannula, M., Vihinen-Ranta, M., Hyttinen, J., Vippola, M., Foi, A., Nymark, S., et al. (2023). Iterative immunostaining combined with expansion microscopy and image processing reveals nanoscopic network organization of nuclear lamina. Mol Biol Cell. 34(9): ee22–09–0448.
分类
生物物理学 > 显微技术
细胞生物学 > 细胞成像 > 超分辨率成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link