- EN - English
- CN - 中文
Detection and Quantification of Programmed Cell Death in Chlamydomonas reinhardtii: The Example of S-Nitrosoglutathione
在莱茵衣藻中检测和定量程序性细胞死亡:以S-硝基谷胱甘肽为例
发布: 2024年08月05日第14卷第15期 DOI: 10.21769/BioProtoc.5043 浏览次数: 1588
评审: Luis Alberto Sánchez VargasAdrian Pascal NievergeltShuhei Ota
Abstract
Chlamydomonas (Chlamydomonas reinhardtii) is a unicellular model alga that has been shown to undergo programmed cell death (PCD) that can be triggered in response to different stresses. We have recently shown that Chlamydomonas is particularly well suited to the study and quantification of PCD. We have shown for the first time that S-nitrosoglutathione (GSNO), a nitric oxide (NO) donor, is able to induce PCD and can be used as a study system in Chlamydomonas. In this article, we provide a simple and robust protocol for quantifying GSNO-induced PCD, which can be adapted to any other treatment. We explain how to detect NO production in the cell following GSNO treatment. We show how PCD can be identified simply by analyzing the degradation profile of genomic DNA. We also provide an easy and reproducible cell death quantification protocol, which makes it possible to follow the course of PCD over time and highlight very fine differences in the number of affected cells between different samples.
Key features
• Use of S-nitrosoglutathione (GSNO) as a means to study programmed cell death (PCD) in Chlamydomonas.
• Discrimination of PCD vs. necrosis.
• In vivo determination of NO production in the cell.
• A simple, robust protocol for PCD quantification.
Keywords: Chlamydomonas reinhardtii (绿藻)Graphical overview
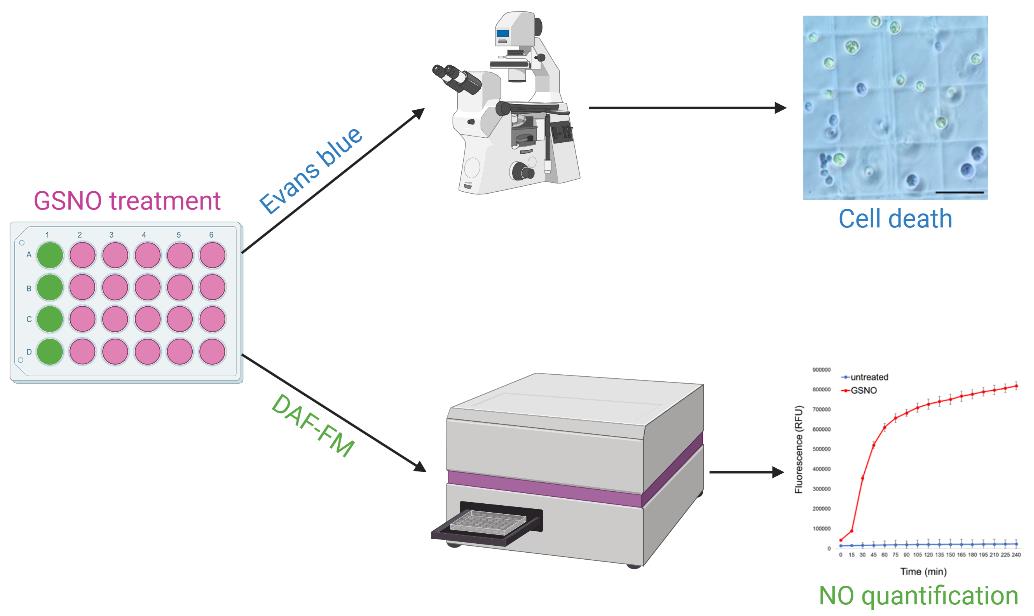
Background
Programmed cell death (PCD) is a crucial process identified in animals in 1972 [1], which plays a role in a wide range of biological processes. Understanding targeted disappearance of cells whose presence is no longer desired is straightforward in multicellular organisms. However, the concept of PCD is more difficult to apply to a unicellular organism, since the death of the cell corresponds to the death of the organism. In recent years, however, it has been shown that PCD does exist in unicellular organisms such as Chlamydomonas, where it was found to be an altruistic mechanism that allows the survival of the population [2,3]. Nitric oxide (NO) has been shown in several studies to be an important molecule for PCD in Chlamydomonas [4,5]; this is why we used the main source of NO in the cell, S-nitrosoglutathione (GSNO), as a system for studying PCD in Chlamydomonas [3]. Several specific criteria can be used to discriminate programmed cell death from necrosis; one of the simplest is to analyze the DNA degradation profile during death [6]. If PCD occurs, DNA migrates as multiples of 180 bp in gel electrophoresis, resulting in a DNA ladder; in the case of necrosis, continuous DNA degradation represented by a smear will be observed [7]. We describe how to analyze the DNA degradation profile in Chlamydomonas, as well as a simple and robust method for calculating the percentage of dead cells in a population. In the case of GSNO-induced PCD, it is important to be able to quantify NO in the cell after treatment; here, we explain how to implement a method for doing so in vivo, using a fluorescent probe. Our protocol outlines the steps to detect and quantify PCD in Chlamydomonas using Evans blue staining, whether you are using GSNO as an inducer or any other treatment.
Materials and reagents
Biological material
We used Chlamydomonas reinhardtii D66 (CC-4425) [8] and CLiP library (CC-4533) [9] strains, but any strain could be used
Reagents
Note: Unless specified otherwise, the reagent can be stored at room temperature.
TAP (tris-acetate-phosphate) solution (Life Technologies, catalog number: T8050)
S-nitrosoglutathione (GSNO) produced in our laboratory but also available for purchase (Sigma, catalog number: N4148), stored at -20 °C
Evans blue powder (Sigma, catalog number: E2129)
Phenol:chloroform:isoamyl (25:24:1) (Sigma, catalog number: 77617), stored at 4 °C
Ethanol 100% (Sigma, catalog number: 32205-M)
Trizma® base (Sigma, catalog number: 93350)
Hydrochloric acid (HCl) (Sigma, catalog number: 258148)
Sodium chloride (NaCl) (Sigma, catalog number: S7653)
Ethylenediaminetetraacetic acid tetrasodium (EDTA) (Sigma, catalog number: E6511)
Sodium dodecyl sulfate (SDS) 20% (Sigma, catalog number: 05030)
Sodium acetate (Sigma, catalog number: S2889)
RNase A, 10 mg/mL (Thermo Scientific, catalog number: EN0531), stored at -20 °C
Agarose (Sigma, catalog number: A9539)
Tris borate EDTA (TBE) buffer (Sigma, catalog number: T4415)
Ethidium bromide solution (Sigma, catalog number: 46067)
GeneRuler 100 bp Plus DNA ladder (Thermo Scientific, catalog number: SM0321), store at 4 °C
DAF-FM diacetate (4-amino-5-methylamino-2’,7’-difluorescein diacetate) (Invitrogen, catalog number: D-23844), store at -20 °C, protected from light
Solutions
50 mM GSNO solution (see Recipes)
DNA extraction buffer (see Recipes)
Sodium acetate 3.3 M (see Recipes)
TBE 0.5× solution (see Recipes)
Evans blue solution (see Recipes)
Recipes
50 mM GSNO solution
Weigh 20 mg of GSNO and dissolve it in 800 µL of TAP medium [10] in a 1.5 mL tube.
Verify the concentration of the 100-fold diluted solution using the molar extinction coefficient of GSNO (922 M-1·cm-1 at 335 nm) [11].
Note: It is better to prepare fresh GSNO for each experiment to avoid its oxidation.
DNA extraction buffer (for 500 mL)
Reagent Final concentration Amount Tris-HCl (1 M, pH 7.5) 200 mM 100 mL NaCl (1 M) 250 mM 125 mL EDTA (0.5 M, pH 8) 25 mM 25 mL SDS (20%) 0.5% 12.5 mL MilliQ water n/a 262.5 mL Total n/a 500 mL Note: It is recommended to autoclave extraction buffer (add SDS after autoclaving) before proceeding to DNA extraction.
Sodium acetate 3.3 M (for 100 mL)
Reagent Final concentration Amount Sodium acetate 3.3 M 27 g MilliQ water n/a 100 mL Total n/a 100 mL Dissolve using a magnetic stirrer and stir bar.
Filter sterilize (0.22 µm).
TBE 0.5× solution
Reagent Final concentration Amount TBE (10×) 0.5× 100 mL MilliQ water n/a 1.9 L Total n/a 2 L Evans blue solution
Reagent Final concentration Amount Evans blue powder 0.5 % 0.5 g TAP n/a 100 mL Total n/a 100 mL Filter sterilize (0.22 µm).
Laboratory supplies
Spectrophotometer semi-micro cuvette (Biosigma, catalog number: BSA002)
24-well plates (Evergreen Labware, Medical Caplugs, catalog number: 222-8044-01F)
Surgical tape (MicroporeTM, catalog number: 1530-0)
Counting chamber Neubauer (Blaubrand, catalog number: BR717805)
Equipment
Spectrophotometer (Implen Nanophotometer)
Microplate reader (ClarioStar Plus, BMG LABTECH)
Orbitron Rotary shaker (Infors)
Olympus BX43 microscope
Thermomixer (Eppendorf, catalog number: 5382000015)
Vortex (Dutsher Vortex Genie 2, catalog number: 079008)
Electrophoresis tanks (Embitec® runOneTM, model: EP-2000)
Camera (Q-IMAGING Micropublisher 3.3 RTV)
NanoDrop One (Thermo Scientific)
GelDoc Go Imaging System (Bio-Rad)
Software and datasets
Image Lab v6.1 for the GelDoc Go Imaging System (Bio-Rad)
Smart control data analysis for ClarioStar Plus (MARS, BMG LabTech)
Procedure
文章信息
稿件历史记录
提交日期: May 13, 2024
接收日期: Jul 2, 2024
在线发布日期: Jul 18, 2024
出版日期: Aug 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Lambert, L. and Danon, A. (2024). Detection and Quantification of Programmed Cell Death in Chlamydomonas reinhardtii: The Example of S-Nitrosoglutathione. Bio-protocol 14(15): e5043. DOI: 10.21769/BioProtoc.5043.
Lambert, L., de Carpentier, F., André, P., Marchand, C. H. and Danon, A. (2024). Type II metacaspase mediates light-dependent programmed cell death in Chlamydomonas reinhardtii.Plant Physiol. 194(4): 2648–2662.
分类
植物科学 > 植物生理学 > 非生物胁迫
细胞生物学 > 细胞信号传导 > 胁迫反应
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













