- EN - English
- CN - 中文
Analysis of Guard Cell Readouts Using Arabidopsis thaliana Isolated Epidermal Peels
利用拟南芥表皮剥离层分析保卫细胞读数
发布: 2024年07月20日第14卷第14期 DOI: 10.21769/BioProtoc.5033 浏览次数: 2310
评审: Wenrong HeVenkatasalam ShanmugabalajiAnonymous reviewer(s)
Abstract
Stomata are pores surrounded by a pair of specialized cells, called guard cells, that play a central role in plant physiology through the regulation of gas exchange between plants and the environment. Guard cells have features like cell-autonomous responses and easily measurable readouts that have turned them into a model system to study signal transduction mechanisms in plants. Here, we provide a detailed protocol to analyze different physiological responses specifically in guard cells. We describe, in detail, the steps and conditions to isolate epidermal peels with tweezers and to analyze i) stomatal aperture in response to different stimuli, ii) cytosolic parameters such as hydrogen peroxide (H2O2), glutathione redox potential (EGSH), and MgATP-2 in vivo dynamics using fluorescent biosensors, and iii) gene expression in guard cell–enriched samples. The importance of this protocol lies in the fact that most living cells on epidermal peels are guard cells, enabling the preparation of guard cell–enriched samples.
Key features
• Isolation of epidermal peels as a monolayer enriched in guard cells.
• Measurement of cytosolic guard cell signaling component dynamics in isolated epidermal peels through fluorescent biosensor analysis.
• Gene expression analysis of guard cell–enriched isolated tissue.
Keywords: Guard cells (保卫细胞)Graphical overview
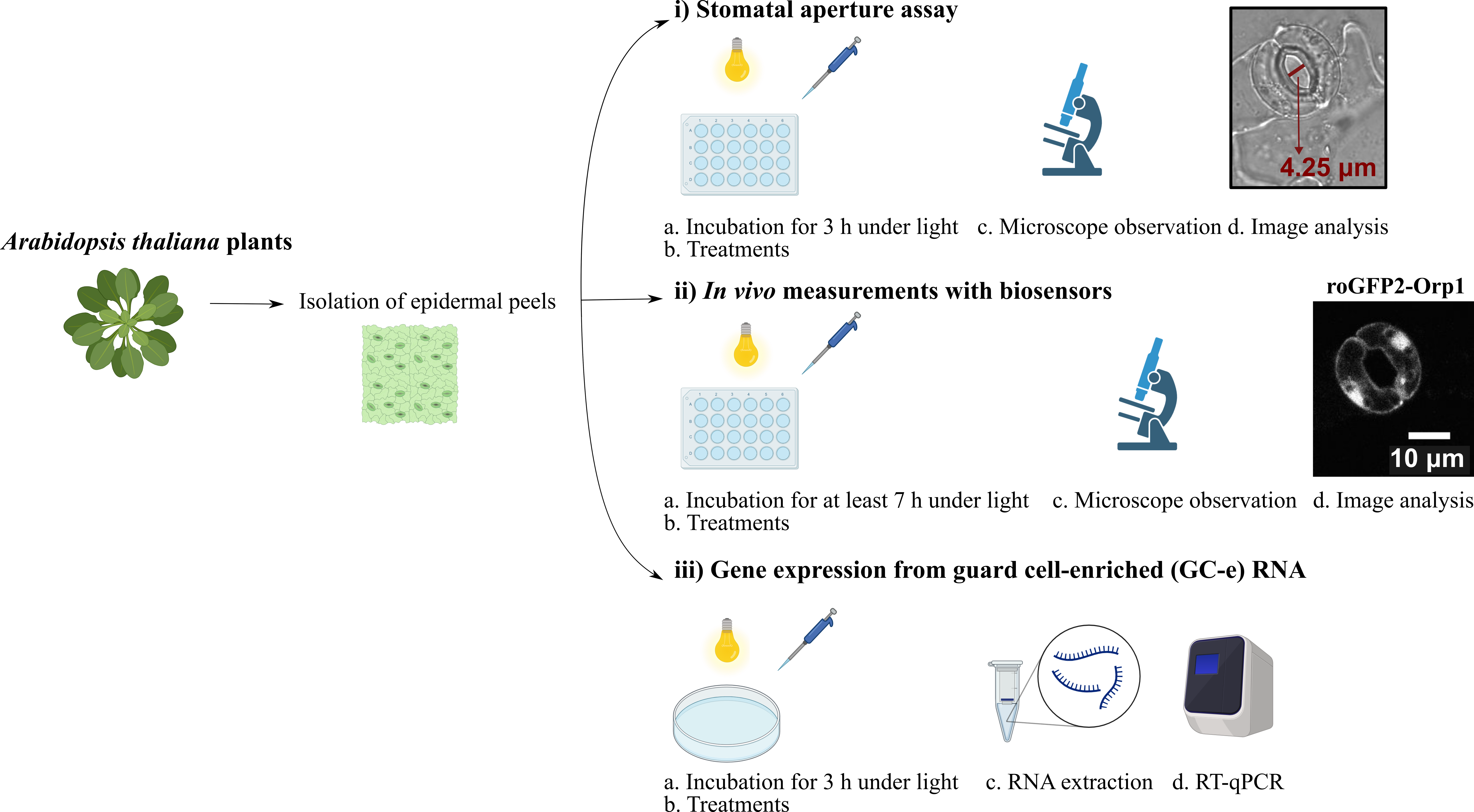
Created with biorender.com
Background
Stomata are microscopic pores located in the epidermis of aerial tissues of most land plants, delimited by specialized cells known as guard cells. Given that the plant epidermis is covered by the cuticle, an external impermeable layer, almost 95% of gas exchange occurs through stomatal pores. Thus, the regulation of the stomatal pore area controls the uptake and release of CO2 and O2 and the maintenance of hydric homeostasis by the regulation of H2O vapor loss, through the transpiration stream [1].
Guard cells continuously sense internal and external cues and integrate them into a complex signaling network that produces modifications in guard cell volume to control the stomatal pore width [2,3]. The isolation of epidermal peels, cellular monolayers mainly formed by functional guard cells, has become a widely used and validated experimental system to study signaling mechanisms in plants [4].
Several protocols have been developed in order to study guard cell physiology [5–7]. Although all of them converge in the isolation of epidermal peels, there are different approaches including the use of adhesive tape [8–10], resin imprints where leaf surface is copied [11–13], blender and filters [14–16], tape and razor blade [17–19], and striping with fine-tip tweezers [20–22].
For the last twenty years, our research group has reproducibly employed fine-tip tweezers to prepare epidermal strips, where almost 90% of living epidermal cells are guard cells (Figure 1A and B) [20,23–27]. Isolation of epidermal peels allowed us to perform stomatal aperture assays and in vivo measurements of endogenously encoded fluorescent proteins (biosensors) that specifically detect hydrogen peroxide (H2O2,), the glutathione redox potential (EGSH), and the biologically relevant portion of adenosine 5'-triphosphate (MgATP2-) [25,27]. In addition, the methodological procedure presented here has been employed to obtain guard cells–enriched RNA (Figure 1B) to analyze transcript levels of different target genes [23,26]. Although peeling produces mechanical stress, evidenced by an increase in reactive oxygen species (ROS) in guard cells, we set up the conditions to recover the strips to basal redox status levels without affecting guard cells’ viability or their physiological responses to different signals [27]. In this protocol, we describe how to prepare isolated epidermal peels excised from the abaxial side of Arabidopsis thaliana leaves and how to perform different experiments using this biological system to study real-time and single-cell physiological processes.
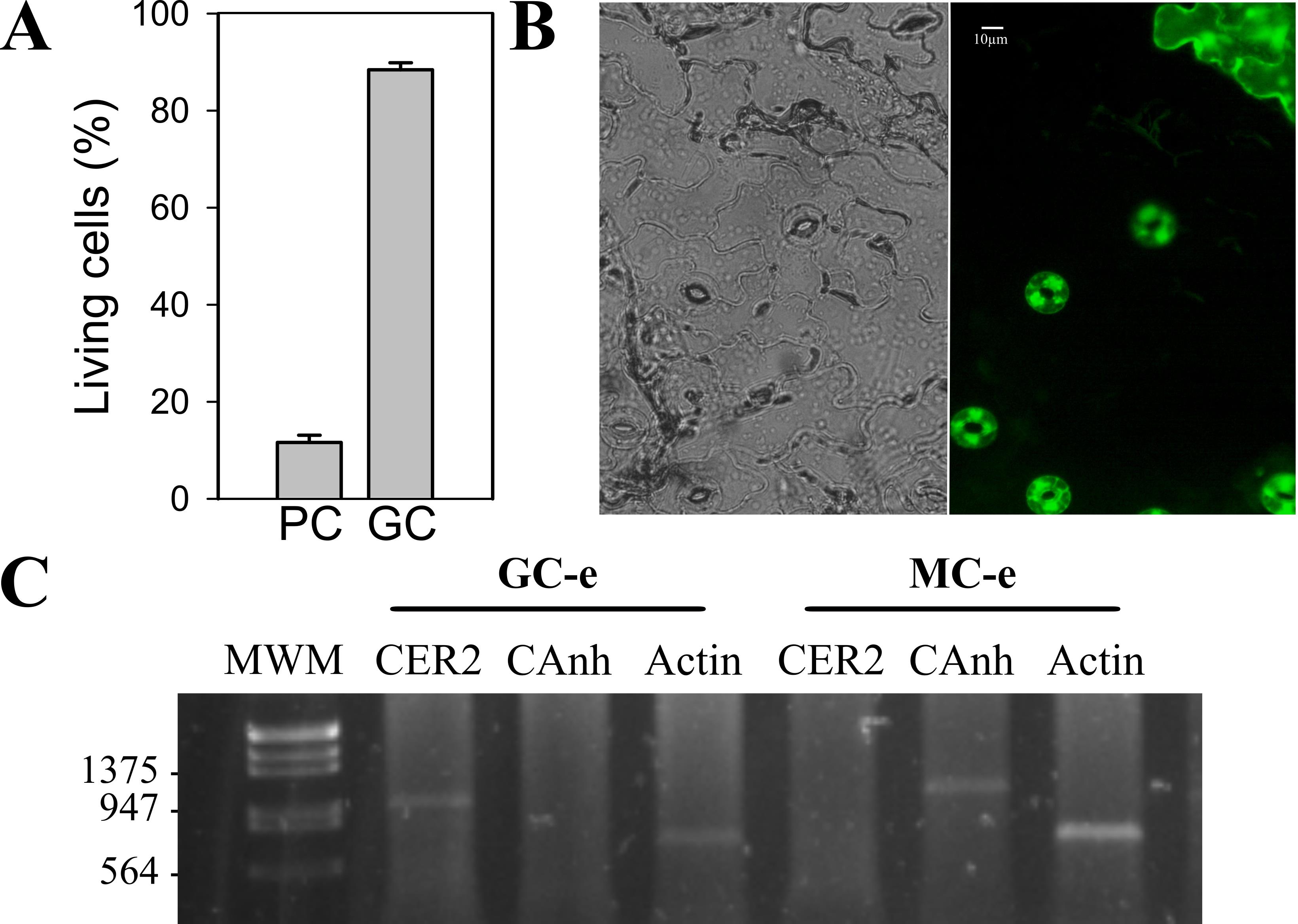
Figure 1. Living cells in epidermal peels. A. Arabidopsis thaliana wild-type ecotype Col-0 epidermal peels were incubated in opening buffer (OB, 5 mM MES pH6.1, 50 mM KCl) under light for 30 min and then loaded with 5 μM of the viability fluorescent dye fluoresce in diacetate (FDA) for 5 min. Peels were washed three times with OB and then mounted on a coverslip, and analyzed by epifluorescence microscopy. Fluorescent cells were quantified, and the total cell/fluorescent cell percentage was calculated for each cell type. PC: pavement cells, GC: guard cells. B. Representative image of an epidermal peel stained with FDA taken with a 40 × objective. Scale bar: 10 μm. C- RT-PCR analysis using guard cell–specific gene ECIREFERUM2 (CER2) (930 bp) and mesophyll cell–specific gene carbonic anhydrase 1 (Canh1) (1,000 bp) was performed in RNA prepared from guard cell–enriched (GC-e) or mesophyll cell–enriched (MC-e) extracts. The amplification of ACTIN (651 bp) transcripts under identical conditions was used as a constitutive expression control. Data from Scuffi et al. [26].
Materials and reagents
Biological materials
4–6 weeks-old wild-type Arabidopsis thaliana (ecotype Col-0) and transgenic Arabidopsis thaliana lines expressing the following biosensors: roGFP2-Orp1 [28], Grx1-roGFP2 [29], and ATeam1.03-nD/nA [30]. See Table 1 to find detailed information regarding the fluorescent biosensor features.
Table 1. Biosensor characteristics
| Biosensor name | roGFP2-Orp1 | Grx1-roGFP2 | ATeam 1.03-nD/nA | |||
| Parameter detected | H2O2 | EGSH | MgATP2- | |||
| Fluorescent protein | EGFP | EGFP | mse CFP, cp173-mVenus | |||
| Promoter | 35S (CaMV) | UBQ10 (from Arabidopsis) | 35S (CaMV) | |||
| Plasmid | pH2GW7 | pBinCM | pH2GW7 | |||
| Selection in plants | Hygromycin | Kanamycin | Hygromycin | |||
| Rationing principle | Dual excitation, Single emission | Dual excitation, Single emission | Single excitation, Dual emission (FRET) | |||
| Emission maximum | 511 nm | 511 nm | 475 nm (mseCFP), 527 nm (cp173-mVenus) | |||
| Microscopy type | Epifluorescence | CLSM | Epifluorescence | CLSM | Epifluorescence | CLSM |
| Excitation wavelength/filter | 450–490 nm, 385–425 nm | 405 nm, 488 nm | 450–490 nm, 385–425 nm | 405 nm, 488 nm | 426–446 nm | 458 nm |
| Emission range | 505–530nm | 505–530 nm(*) | 505–530 nm | 505–530 nm(*) | 467–499 nm, 528.5–555.5 nm(**) | 465–500 nm, 526–561 nm(*) |
| References | (15) | (14), (22) | (15) | (11): (22) | (15) | (3) |
(*) A 650–695 nm filter can be used for chlorophyll fluorescence collection.
(**) Use a dichroic 510–440 nm mirror for simultaneous acquisitions.
CSLM: Confocal scanning laser microscopy.
Reagents
Soil (Antoniucci, https://viveroantoniucci.mitiendanube.com/productos/tierra-negra/)
Vermiculite (Terrafertil, https://www.terrafertil.com/productos_jardin/acondicionadores_vermiculita.html)
Perlite (Terrafertil, https://www.terrafertil.com/productos_jardin/acondicionadores_vermiculita.html)
2-(N-Morpholino) ethane sulfonic acid (MES) (Sigma, CAS: 4432-31-9)
Potassium chloride (KCl) (Sigma, CAS: 7447-40-7)
Potassium hydroxide (KOH) (Sigma, CAS: 1310-58-3)
Trizol (Invitrogen, catalognumber: 15596026)
Opening buffer (see Recipes)
Opening buffer
Reagent stock Final concentration Amount MES (50 mM, pH 6.1)* 5 mM 10 mL KCl (1M) 50 mM 5 mL H2O n/a 85 mL Total n/a 100 mL (*) MES is adjusted to pH with KOH
Laboratory supplies
Precision tweezers, Dumont #5 (Fine Science Tool, catalog number: 11251-10)
Scalpel n° 4 (KLS Martin, catalog number: 10-100-04)
1 mL syringe with needle with the tip bent, n° 27 g (Bremen Seiseme, catalog number: 7791914003029)
Pipette tips [Deltalab, catalog number: 327-34 (2-200 μL), 200070 (1,000 μL)]
Pipettes [Gilson, Pipetman, catalog number: F144056M (P20), F144058M (P200), F144059M (P1000)]
24-well plate (BIOFIL, catalog number: TCP001024)
Petri dish 55 × 14 mm (Deltalab, catalog number: 200201)
1.5 mL tubes (Deltalab, catalog number: 200400)
Microscope slides (Deltalab, catalog number: D100010)
Coverslips 18 × 18 mm nº 1 (Deltalab, catalog number: D101818)
Neubauer chamber (BOECO Germany, catalog number: BOE01)
Mortar (Günther Argentina, catalog number: 14660) and pestle (Günther Argentina, catalog number: 154200)
Liquid nitrogen
Equipment
Brightfield microscope (Olympus, model: CKX53) coupled to a digital camera (AmScope, model: MU1000) and 40 × objective (LUC Plan FLN, numerical aperture: 0.6)
Confocal laser scanning microscope (CLSM) (Carl Zeiss Microscopy, model: LSM980) with a 40 × water immersion objective (C-Apochromat, 1.2 numerical aperture)
Epifluorescence microscope (Nikon, model: Ti-E) coupled to a double digital camera (Hamamatsu, model: ORCA-D2 Dual CCD) and 60 × oil immersion objective (CFI Plan APO Lambda, 1.4 numerical aperture)
Software and datasets
NIS-Elements AR (https://www.microscope.healthcare.nikon.com/products/software/nis-elements/nis-elements-advanced-research)
Fiji analysis software (ImageJ), National Institutes of Health, Version 1.53t [31]
Procedure
文章信息
稿件历史记录
提交日期: Mar 4, 2024
接收日期: Jun 16, 2024
在线发布日期: Jul 3, 2024
出版日期: Jul 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Pantaleno, R., Schiel, P., García-Mata, C. and Scuffi, D. (2024). Analysis of Guard Cell Readouts Using Arabidopsis thaliana Isolated Epidermal Peels. Bio-protocol 14(14): e5033. DOI: 10.21769/BioProtoc.5033.
- Scuffi, D., Nietzel, T., DiFino, L. M., Meyer, A. J., Lamattina, L., Schwarzländer, M., Laxalt, A. M. and García-Mata, C. (2018). Hydrogen Sulfide Increases Production of NADPH Oxidase-Dependent Hydrogen Peroxide and Phospholipase D-Derived Phosphatidic Acid in Guard Cell Signaling. Plant Physiol. 176 (3): 2532–2542. https://doi.org/10.1104/pp.17.01636
分类
植物科学 > 植物细胞生物学 > 细胞成像
细胞生物学 > 细胞信号传导 > 第二信使
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













