- EN - English
- CN - 中文
Versatile Cloning Strategy for Efficient Multigene Editing in Arabidopsis
拟南芥高效多基因编辑的多用途克隆策略
发布: 2024年07月05日第14卷第13期 DOI: 10.21769/BioProtoc.5029 浏览次数: 2220
评审: Raniki KumariAnonymous reviewer(s)
Abstract
CRISPR-Cas9 technology has become an essential tool for plant genome editing. Recent advancements have significantly improved the ability to target multiple genes simultaneously within the same genetic background through various strategies. Additionally, there has been significant progress in developing methods for inducible or tissue-specific editing. These advancements offer numerous possibilities for tailored genome modifications. Building upon existing research, we have developed an optimized and modular strategy allowing the targeting of several genes simultaneously in combination with the synchronized expression of the Cas9 endonuclease in the egg cell. This system allows significant editing efficiency while avoiding mosaicism. In addition, the versatile system we propose allows adaptation to inducible and/or tissue-specific edition according to the promoter chosen to drive the expression of the Cas9 gene. Here, we describe a step-by-step protocol for generating the binary vector necessary for establishing Arabidopsis edited lines using a versatile cloning strategy that combines Gateway® and Golden Gate technologies. We describe a versatile system that allows the cloning of as many guides as needed to target DNA, which can be multiplexed into a polycistronic gene and combined in the same construct with sequences for the expression of the Cas9 endonuclease. The expression of Cas9 is controlled by selecting from among a collection of promoters, including constitutive, inducible, ubiquitous, or tissue-specific promoters. Only one vector containing the polycistronic gene (tRNA-sgRNA) needs to be constructed. For that, sgRNA (composed of protospacers chosen to target the gene of interest and sgRNA scaffold) is cloned in tandem with the pre-tRNA sequence. Then, a single recombination reaction is required to assemble the promoter, the zCas9 coding sequence, and the tRNA-gRNA polycistronic gene. Each element is cloned in an entry vector and finally assembled according to the Multisite Gateway® Technology. Here, we detail the process to express zCas9 under the control of egg cell promoter fused to enhancer sequence (EC1.2en-EC1.1p) and to simultaneously target two multiple C2 domains and transmembrane region protein genes (MCTP3 and MCTP4, respectively at3g57880 and at1g51570), using one or two sgRNA per gene.
Key features
• A simple method for Arabidopsis edited lines establishment using CRISPR-Cas9 technology
• Versatile cloning strategy combining various technologies for convenient cloning (Gateway®, Golden Gate)
• Multigene targeting with high efficiency
Keywords: Plant genome editing (植物基因组编辑)Background
CRISPR-Cas9 technology, which serves as a powerful genome editing tool, is based on the bacterial RNA-guided CRISPR-Cas9 system [1]. The editing process involves two main actors: the Cas9 nuclease and a unique guide RNA (sgRNA) that will direct the Cas9 protein to the DNA target for genome editing. Plants, like other organisms, need these elements to form the CRISPR-Cas9 complex, which will be directed to the targeted sequence depending on the crRNA part of the sgRNA [2]. Plant transformation with binary vectors containing sequences for Cas9 expression and sgRNA synthesis remains one of the primary and widely used methods for the production of these actors in cells. Numerous vectors and editing strategies have been developed, such as strategies to target multiple genes simultaneously through sgRNA multiplexing. This approach addresses challenges such as gene redundancy and enhancing editing efficiency, eliminating the need for time-consuming multiple crossings [3–6]. Other ways of improvement using various promoters have been explored to optimize the expression of the Cas9, especially in the context of multitargeting but also to avoid mosaic mutations. Indeed, while promoter UBQ10 enhances mutation efficiency in comparison to the 35S promoter, ubiquitous promoters driving the expression of Cas9 could lead to mosaic mutation patterns [7]. To fix this problem, egg cell–specific promoters like EC1.2, embryo sac, embryo, and endosperm, and pollen-specific promoters like YAO or NUC1 can be used. In these conditions, Cas9 expression induces homozygous or biallelic mutants in the early generations removing the problem of mosaicism [8–10]. Taking advantage of previously published works, we chose efficient elements and strategies and merged them to develop a very versatile cloning system that combines MultiSite Gateway® [11] and Golden Gate [12] technologies.
In this protocol, we describe the step-by-step procedure for generating the binary vector that contains the endogenous tRNA-processing system, previously described to boost the targeting and multiplex editing capability of the CRISPR/Cas9 system [4], and the egg cell–specific promoter EC1.2 associated to EC1.1 enhancer to drive the expression of Cas9 [8] (Figure 1). For precise genome editing, this protocol can be readily adjusted to accommodate various designs of binary vectors. This includes the utilization of different tissue-specific promoters, inducible promoters, or alternative nucleases such as SaCas9, Cas12a, or zCas9i, which can enhance both the efficiency and specificity of editing [13–17].
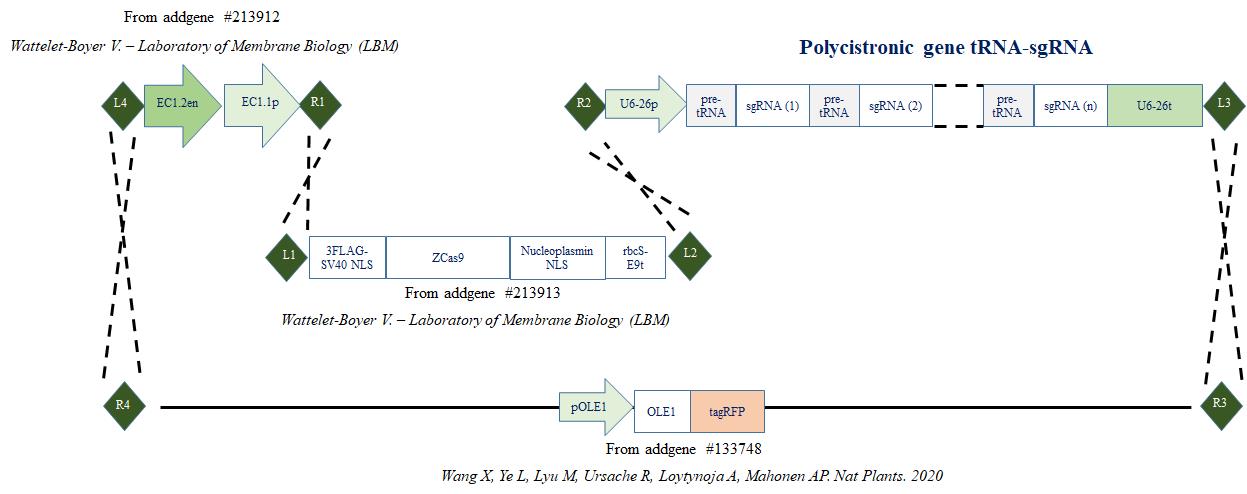
Figure 1. Cloning strategy. Cloning strategy to express zCas9 under the control of egg cell promoter (EC1.1p) fused to enhancer sequence (EC1.2en) and to target DNA using several sgRNA cloned in the form of a polycistronic gene. (R1, R2, R3, R4, L1, L2, L3 and L4 represent recombination sites.)
Materials and reagents
Biological materials
pHEE401E plasmid (Addgene, catalog number: 71287)
pGTR plasmid (Addgene, catalog number: 63143)
pFRm43GW plasmid (Addgene, catalog number: 133748)
p4P1R_EC1.2en-EC1.1p plasmid (Addgene, catalog number: 213912)
pDONR221_zCas9 plasmid (Addgene, catalog number: 213913)
pDONRTM P2R-P3 (Thermo Fisher Scientific, catalog number: 12537-023)
One ShotTM TOP10 chemically competent E. coli (Thermo Fisher Scientific, catalog number: C404010)
List of primers common to all strategies
Order common and specific primers according to the following features:
Synthesis scale: 0.05 μmol; purification: cartridge; format: in solution (water); concentration: 100 μM.
(Supplier: Merck, but could be another one)
Fw-Primer 1: 5′- GGGGACAGCTTTCTTGTACAAAGTGGAACGACTTGCCTTCCGCACAATAC - 3’
Rev-Primer 2: 5′- atggtctcaTGTTAATCACTACTTCGACTCTAG - 3′
Fw-Primer 3: 5′- taggtctccAACAAAGCACCAGTGGTCTAGTGG - 3′
Rev-Primer 12: 5′- atggtctcaAAGCACCGACTCGGTGCCACTTTTTC - 3′
Fw-Primer 13: 5′- taggtctccGCTTTTTTTTGCAAAATTTTCCAGTCG - 3′
Rev-Primer 14: 5′- GGGGACAACTTTGTATAATAAAGTTGATATTGGTTTATCTCATCGGAAC - 3′
M13-Fw (–20): 5′- GTAAAACGACGGCCAG - 3′
M13-Rev: 5′- CAGGAAACAGCTATGAC - 3′
Reagents
Enzymes for molecular biology
Q5® high-fidelity DNA polymerase (NEB, catalog number: M0491)
OneTaq 2× Master Mix with standard buffer (NEB, catalog number: M0482)
BsaI-HF restriction enzyme (NEB, catalog number: R3733)
T4 DNA ligase (Thermo Fisher Scientific, catalog number: 15224025)
GatewayTM BP ClonaseTM II (Thermo Fisher Scientific, catalog number: 11789020)
GatewayTM LR ClonaseTM II (Thermo Fisher Scientific, catalog number: 11791020)
Reagents for molecular biology
Cutsmart buffer (NEB, catalog number: B6004)
dNTP solution MIX 10 mM (NEB, catalog number: N0447)
TAE (Tris-Acetate-EDTA buffer) (Euromedex, catalog number: EU0202)
SYBRTM Safe DNA gel stain (Thermo Fisher Scientific, catalog number: S33102)
Agarose (Euromedex, catalog number: LE-8200)
DNase-free water (autoclaved at 110 °C for 30 min)
Reagents for microbiology
LB Miller (Euromedex, catalog number: AE-0103)
Bacteriological agar (Euromedex, catalog number: 1330)
Kanamycin bisulfate (Euromedex, catalog number: EU0420)
Spectinomycin dihydrochloride pentahydrate (Merck, catalog number: S4014)
Kit for molecular biology
Monarch® PCR & DNA Cleanup kit (NEB, catalog number: T1030)
NucleoSpin® Plasmid kit (Macherey Nagel, catalog number: 740588)
Kanamycin bisulfate stock solution (25 mg/mL) (see Recipes)
Spectinomycin dihydrochloride pentahydrate stock solution (50 mg/mL) (see Recipes)
LB Miller media (see Recipes)
Kanamycin bisulfate stock solution (25 mg/mL)
Reagent Final concentration Amount Kanamycin bisulfate 25 mg/mL 0.25 g ddH2O n/a 10 mL Total n/a 10 mL Filter with a 0.22 μm filter, aliquot by 1 mL, and store at -20 °C.
Spectinomycin dihydrochloride pentahydrate stock solution (50 mg/mL)
Reagent Final concentration Amount Spectinomycin dihydrochloride pentahydrate 50 mg/mL 0.5 g ddH2O n/a 10 mL Total n/a 10 mL Filter with a 0.22 μm filter, aliquot by 1 mL, and store at -20 °C.
LB Miller media
Reagent Final concentration Amount LB Miller 25 g/L 25 g Bacteriological agar 20 g/L 20 g ddH2O n/a 1,000 mL Total n/a 1,000 mL Autoclave at 110 °C for 30 min Wait until the temperature of the media is at approximately 60 °C and add the antibiotic (final concentration: kanamycin bisulfate 25 μg/mL or spectinomycin dihydrochloride pentahydrate 50 μg/mL).
Laboratory supplies
Reaction tubes, 0.5 mL, PP (SARSTEDT, catalog number: 72699)
Reaction tubes, 1.5 mL, PP (SARSTEDT, catalog number: 72690001)
Petri dishes, Ø 90 mm (VWR, catalog number: 391-0556)
Wooden toothpicks length 80 mm (DUTSCHER, catalog number: 505802)
Glass beads, Ø 5 mm (DUTSCHER, catalog number: 068503)
10/20 μL XL graduated TipOne® tips (STARLAB, catalog number: S1110-3700-C)
200 μL UltraPoint® graduated TipOne® tips (STARLAB, catalog number: S1113-1700-C)
1250 μL XL graduated TipOne® tips (STARLAB, catalog number: S1112-1720-C)
Microtubes 0.2 mL + flat caps (DUTSCHER, catalog number: 010208)
Equipment
Thermocycler (BIORAD, C1000 Touch Thermal Cycler, catalog number: 1851148)
Complete Vortex Genie 2 (DOMINIQUE DUTSCHER, catalog number: 079008)
Electrophoresis chamber Mupid ONE (DOMINIQUE DUTSCHER, catalog number: 088900)
Water bath (VWR, catalog number: IKAA20004382)
Shaking incubator, digital, benchtop, ES-20 (VWR, catalog number: 444-0936)
Oven (DOMINIQUE DUTSCHER, catalog number: 485134)
NanoDrop 2000 (THERMOFISHER SCIENTIFIC, catalog number: ND-2000)
Software and datasets
Web application tool CRISPR-P (v1.0, 2014) (http://crispr.hzau.edu.cn/CRISPR/) [18]
Procedure
文章信息
稿件历史记录
提交日期: Apr 4, 2024
接收日期: Jun 11, 2024
在线发布日期: Jul 2, 2024
出版日期: Jul 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Li, Z. P., Huard, J., Bayer, E. and Wattelet-Boyer, V. (2024). Versatile Cloning Strategy for Efficient Multigene Editing in Arabidopsis. Bio-protocol 14(13): e5029. DOI: 10.21769/BioProtoc.5029.
分类
植物科学 > 植物分子生物学 > 遗传分析
分子生物学 > DNA > 基因表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











