- EN - English
- CN - 中文
A Single-step Generation of AlissAID-based Conditional Knockdown Strains Using Nanobody that Targets GFP or mCherry in Budding Yeast
利用靶向GFP或mCherry的纳米抗体在出芽酵母中一步生成AlissAID条件性敲降菌株
发布: 2024年06月20日第14卷第12期 DOI: 10.21769/BioProtoc.5019 浏览次数: 2056
评审: Lucy XieOlga SinJoyce ChiuAnonymous reviewer(s)
Abstract
The Auxin-inducible degron (AID) system is a genetic tool that induces rapid target protein depletion in an auxin-dependent manner. Recently, two advanced AID systems—the super-sensitive AID and AID 2—were developed using an improved pair of synthetic auxins and mutated TIR1 proteins. In these AID systems, a nanomolar concentration of synthetic auxins is sufficient as a degradation inducer for target proteins. However, despite these advancements, AID systems still require the fusion of an AID tag to the target protein for degradation, potentially affecting its function and stability. To address this limitation, we developed an affinity linker–based super-sensitive AID (AlissAID) system using a single peptide antibody known as a nanobody. In this system, the degradation of GFP- or mCherry-tagged target proteins is induced in a synthetic auxin (5-Ad-IAA)–dependent manner. Here, we introduce a simple method for generating AlissAID strains targeting GFP or mCherry fusion proteins in budding yeasts.
Key features
• AlissAID system enables efficient degradation of the GFP or mCherry fusion proteins in a 5-Ad-IAA–depending manner.
• Transforming the pAlissAID plasmids into strains with GFP- or mCherry- tagged proteins.
Keywords: Auxin-inducible degron (AID) (生长素诱导降解系统 (AID))Background
Targeted protein degradation (TPD) is a powerful tool for investigating protein function by rapidly depleting the target proteins in cells. Auxin-inducible degron (AID) is a TPD system that enables auxin-dependent AID-tagged target protein degradation [1]. This technique has been widely used in various eukaryotic species, including yeast, Drosophila, Caenorhabditis elegans, and vertebrate cells [2–5]. In this system, expression of the AID-tagged target protein and the auxin receptor Oriza sative TIR1 (OsTIR1) are necessary for target degradation. The OsTIR1 interacts with the AID degron, an interaction that is stabilized by auxin. It further recruits the E3 ubiquitin ligase Skp1-Cul1-F-box (SCF), which leads to the ubiquitination and degradation of target proteins.
The conventional AID system requires a concentration of 100 µM or higher of auxin for the target protein degradation. However, such a high dose of auxin may lead to cytotoxicity [6]. To address this issue, two alternative systems, super-sensitive AID (ssAID) [7] and AID2 [8] have been developed using the bump and hole technique. The ssAID system uses a high-affinity pair of synthetic auxin, 5-Adamantyl-IAA (5-Ad-IAA), and a modified auxin receptor, OsTIR1F74A. This advanced system enables the degradation of AID-tagged target proteins at nanomolar concentrations of 5-Ad-IAA.
Recently, we developed an affinity-linker–based supersensitive AID (AlissAID) system using a single polypeptide antibody known as a nanobody (Figure 1) [9]. In this system, the addition of 5-Ad-IAA induces the degradation of GFP- or mCherry-tagged proteins by stabilizing the interaction between OsTIR1F74A and the minimized AID-tagged nanobody (mAID-Nb). [9]. The binding of OsTIR1F74A and mAID-tag depends on the concentration of 5-Ad-IAA. This advancement enables the use of GFP or mCherry proteins as degradation tags instead of the conventional AID tags in the AlissAID system.
GFP and mCherry are well-known fluorescent tags used in various eukaryotic cells. In budding yeast, GFP fusion with endogenous proteins is easily achievable through genetic manipulation. Furthermore, the GFP tag can be employed in a Yeast GFP Clone collection [10], with a GFP-tagged Open Reading Frame (ORF) at its chromosomal locus, containing 75% of the yeast proteome. Here, we describe a protocol for easy AlissAID strain generation from GFP- or mCherry-tagged strains in budding yeast.
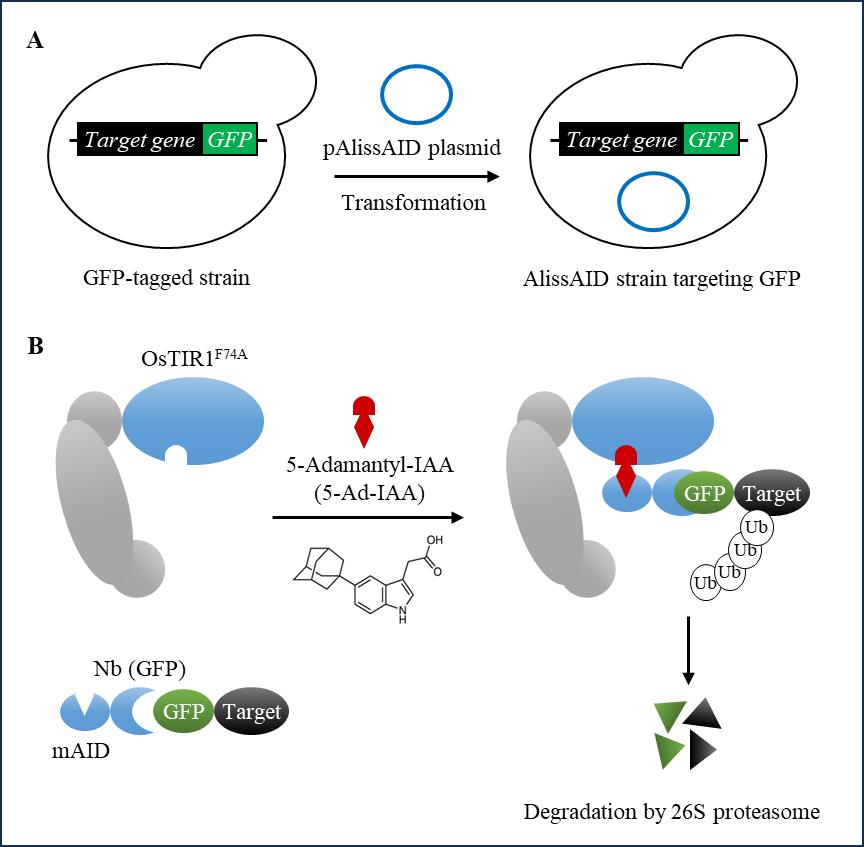
Figure 1. Schematic illustration of generating AlissAID strains from GFP-tagged strains. (A) To generate the AlissAID strain, the pAlissAID anti-GFP plasmid is transformed into a GFP-tagged strain. (B) OsTIR1F74A and mAID-Nb (GFP) are constitutively expressed from the pAlissAID anti-GFP plasmid. The expressed OsTIR1F74A forms the SCF-OsTIR1F74A complex and then polyubiquitinates the GFP fusion protein in a 5-Ad-IAA dependent manner.
Materials and reagents
Biological materials
AlissAID plasmids targeting GFP fusion proteins
Note: These plasmids are available from National BioResource Project (NBRP)– Yeast
https://yeast.nig.ac.jp/yeast/top.xhtml
pAlissAID anti-GFP_313 (HIS3 marker) (NBRP, catalog number: BYP10393)
Vector map: https://yeast.nig.ac.jp/yeast/pdf/byp/BYP10393.pdf
pAlissAID anti-GFP_314 (TRP1 marker) (NBRP, catalog number: BYP10394)
Vector map: https://yeast.nig.ac.jp/yeast/pdf/byp/BYP10394.pdf
pAlissAID anti-GFP_315 (LEU2 marker) (NBRP, catalog number: BYP10395)
Vector map: https://yeast.nig.ac.jp/yeast/pdf/byp/BYP10395.pdf
pAlissAID anti-GFP_316 (URA3 marker) (NBRP, catalog number: BYP10392)
Vector map: https://yeast.nig.ac.jp/yeast/pdf/byp/BYP10392.pdf
AlissAID plasmids targeting for mCherry fusion proteins
pAlissAID anti-mCherry_313 (HIS3 marker) (NBRP, catalog number: BYP10396)
Vector map: https://yeast.nig.ac.jp/yeast/pdf/byp/BYP10396.pdf
pAlissAID anti-mCherry_314 (TRP1 marker) (NBRP, catalog number: BYP10397)
Vector map: https://yeast.nig.ac.jp/yeast/pdf/byp/BYP10397.pdf
pAlissAID anti-mCherry_315 (LEU2 marker) (NBRP, catalog number: BYP10398)
Vector map: https://yeast.nig.ac.jp/yeast/pdf/byp/BYP10398.pdf
pAlissAID anti-mCherry_316 (URA3 marker) (NBRP, catalog number: BYP10399)
Vector map: https://yeast.nig.ac.jp/yeast/pdf/byp/BYP10399.pdf
Budding yeast strains (commonly used strains are useable, BY4741 W303-1a, etc…)
Antibodies (anti-OsTIR1 (MBL, catalog number: PD048), anti-Pgk1 (our Laboratory), anti-AIDtag (gifted from Prof. Karim Labib)
Lithium acetate (LioAc) (Wako, catalog number: 127-01545)
Salmon sperm DNA (ssDNA) (Wako; catalog number: 043-31381)
Poly-ethylene glycol 4,000 (PEG) (Wako, catalog number: 162-09115)
Dimethyl sulfoxide (DMSO) (Wako, catalog number: 043-07216)
5-Adamantyl-IAA (5-Ad-IAA) (Tokyo Chemical Industry (catalog number: A3390).
D-Glucose (Wako, catalog number: 045-31167)
Extract yeast dried (Nacalai Tesque, catalog number: 15838-45)
Polypeptone (Wako, catalog number: 398-02117)
Adenine hydrochloride (Biosynth, catalog number: FA02944)
Yeast nitrogen base without amino acids (ForMedium, catalog number: CYN0410)
SC Quadruple Drop Out: -His, -Leu, -Trp, -Ura (ForMedium, catalog number: DSCK1027)
Agar (Nacalai Tesque, catalog number: 01028-85)
YPD medium (see Recipes)
SD medium (see Recipes)
Amino acids and Uracil solution (see Recipes)
LiOAc solution (0.1 M, 1 M) (see Recipes)
Single-strand DNA (ssDNA; 2.0 mg/mL) (see Recipes)
PEG (50% w/v) (see Recipes)
5-Ad-IAA (see Recipes)
YPD medium
Reagent Final concentration Amount D-glucose 2% 20 g Extract yeast dried 1% 10 g peptone 2% 20 g Adenine hydrochloride 0.01% 100 mg MILLI-Q Total 1,000 mL SD medium
Reagent Final concentration Amount D-glucose 2% 20 g Adenine hydrochloride 0.01% 100 mg Yeast nitrogen base without amino acids 0.69% 6.9 g SC Quadruple Drop Out: -His, -Leu, -Trp, -Ura 0.06% 0.6 g 5 M NaOH 5 mM 1 mL agar (for plate) 2% 20 g Amino acids and Uracil solution 10 mL MILLI-Q Total 1,000 mL For auxotrophic selection, supplement appropriate amino acids and Uracil.
Amino acids and Uracil solution
Histidine solution 10 g/L
Leucine solution 12 g/L
Tryptophan solution 10 g/L
Uracil solution 2 g/L
LiOAc solution (0.1 M, 1 M)
Dissolve in MILLI-Q at the prescribed concentration and autoclave.
Single-strand DNA (ssDNA; 2.0 mg/mL)
Dissolve in MILLI-Q at a concentration of 2.0 mg/mL and autoclave.
PEG (50% w/v)
Dissolve in MILLI-Q water at 50% (w/v) and autoclave. The transformation efficiency will decrease if used for a long time; therefore, approximately 10 samples should be prepared at a time.
5-Ad-IAA
Dissolve in DMSO at a concentration of 5 mM and store at -30 °C. When it is used for liquid medium, add 1/1,000 of the amount of 5 mM 5-Ad-IAA to the medium. When it is used for plate media, autoclaved media should be cooled to approximately 50 °C before adding 5-Ad-IAA. 5-Ad-IAA containing plates can be stocked at 4°C, hidden from direct light, for at least one month.
Laboratory supplies
Laboratory disposables:
1.5 mL tubes (Watson, catalog number: 131-7155C)
15 mL tubes (Greiner, catalog number: 188 271- 013)
50 mL tubes (Greiner, catalog number: 227 261)
1000 μL tips (Watson, catalog number: 110-706C)
200 μL tips (Watson, catalog number: 110-705C)
Petri dishes (STAR, catalog number: RSU-SD9015-2)
Equipment
Cool incubator (As one, catalog number: A1201)
Shaking incubator (N-BIOTEK, catalog number: NB-205L)
Heat block (WAKENYAKU, catalog number: WKN-9626)
Vortex (Scientific Industries, Inc., catalog number: SI-0286)
Centrifuge (Eppendorf, catalog number: 5420000237
Microscope (AxioObserver Z1 (Carl Zeiss, Oberkochen, Germany) equipped with a CCD camera (AxioCam MRm; Carl Zeiss))
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Ogawa, Y., Ueda, T. P., Obara, K., Nishimura, K. and Kamura, T. (2024). A Single-step Generation of AlissAID-based Conditional Knockdown Strains Using Nanobody that Targets GFP or mCherry in Budding Yeast. Bio-protocol 14(12): e5019. DOI: 10.21769/BioProtoc.5019.
分类
微生物学 > 微生物遗传学 > 质粒
细胞生物学 > 细胞工程
生物化学 > 蛋白质 > 降解
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











