- EN - English
- CN - 中文
CD8α-CI-M6PR Particle Motility Assay to Study the Retrograde Motion of CI-M6PR Receptors in Cultured Living Cells
CD8α-CI-M6PR颗粒运动测定研究培养活细胞中CI-M6PR受体的逆行运动
发布: 2024年05月05日第14卷第9期 DOI: 10.21769/BioProtoc.4979 浏览次数: 1659
评审: Chiara AmbrogioDevashish DWIVEDI
Abstract
The cation-independent mannose 6-phosphate receptors (CI-M6PR) bind newly synthesized mannose 6-phosphate (Man-6-P)-tagged enzymes in the Golgi and transport them to late endosomes/lysosomes, providing them with degradative functions. Following the cargo delivery, empty receptors are recycled via early/recycling endosomes back to the trans-Golgi network (TGN) retrogradely in a dynein-dependent motion. One of the most widely used methods for studying the retrograde trafficking of CI-M6PR involves employing the CD8α-CI-M6PR chimera. This chimera, comprising a CD8 ectodomain fused with the cytoplasmic tail of the CI-M6PR receptor, allows for labeling at the plasma membrane, followed by trafficking only in a retrograde direction. Previous studies utilizing the CD8α-CI-M6PR chimera have focused mainly on colocalization studies with various endocytic markers under steady-state conditions. This protocol extends the application of the CD8α-CI-M6PR chimera to live cell imaging, followed by a quantitative analysis of its motion towards the Golgi. Additionally, we present an approach to quantify parameters such as speed and track lengths associated with the motility of CD8α-CI-M6PR endosomes using the Fiji plugin TrackMate.
Key features
• This assay is adapted from the methodology by Prof. Matthew Seaman for studying the retrograde trafficking of CI-M6PR by expressing CD8α-CI-M6PR chimera in HeLa cells.
• The experiments include live-cell imaging of surface-labeled CD8α-CI-M6PR molecules, followed by a chase in cells.
• Allows the monitoring of real-time motion of CD8α-CI-M6PR endosomes and facilitates calculation of kinetic parameters associated with endosome trajectories, e.g., speed and distance (run lengths).
Keywords: Trafficking (运输)Background
At steady state, the cation-independent mannose 6-phosphate receptors (CI-M6PR) are primarily localized to the trans-Golgi network (TGN), with a perinuclear endosomal population and a minor fraction on the plasma membrane. Post cargo delivery to endolysosomal compartments, CI-M6PR receptors undergo trafficking from all the cellular locations to early/recycling endosomes; subsequently, they are retrieved back to TGN in a dynein-dependent manner. This endosome-to-TGN retrieval of CI-M6PR is crucial for maintaining lysosomal activity and is regulated by various proteins involved in sorting and packaging into carrier vesicles followed by their motility towards TGN. Dysregulation in CI-M6PR receptor trafficking is linked to neurodegenerative and lysosomal storage disorders such as Batten's disease and Parkinson's disease [1,2]. Initial studies on CI-M6PR trafficking either required radiolabeling newly synthesized CI-M6PR and tracking their journey from Golgi or predominantly relied on measuring the redistribution of CI-M6PR receptors from TGN to other cellular compartments [3]. However, these methodologies only provide information on the localization of the cargo upon its impaired trafficking without revealing the exact direction of trafficking defect. Recently, several studies have employed the CD8α-CI-M6PR chimera (developed by Matthew Seaman; [4]) to study retrograde trafficking of CI-M6PR, involving colocalization of CD8α-CI-M6PR with different endocytic markers at steady-state. This chimera, a fusion of the ectodomain of CD8 and the cytoplasmic tail of CI-M6PR, allows trafficking only in the retrograde direction towards the Golgi [5]. Here, we present a protocol that extends the use of CD8α-CI-M6PR chimera in a live-cell imaging setup, where surface receptors are labeled and then chased in cells as they move towards the Golgi (Figure 1). We also provide an approach to perform the quantitative analysis of CD8α-CI-M6PR motion as they move from early/recycling endosome to Golgi, including the calculation of kinetic parameters such as speed and distance associated with their motion.
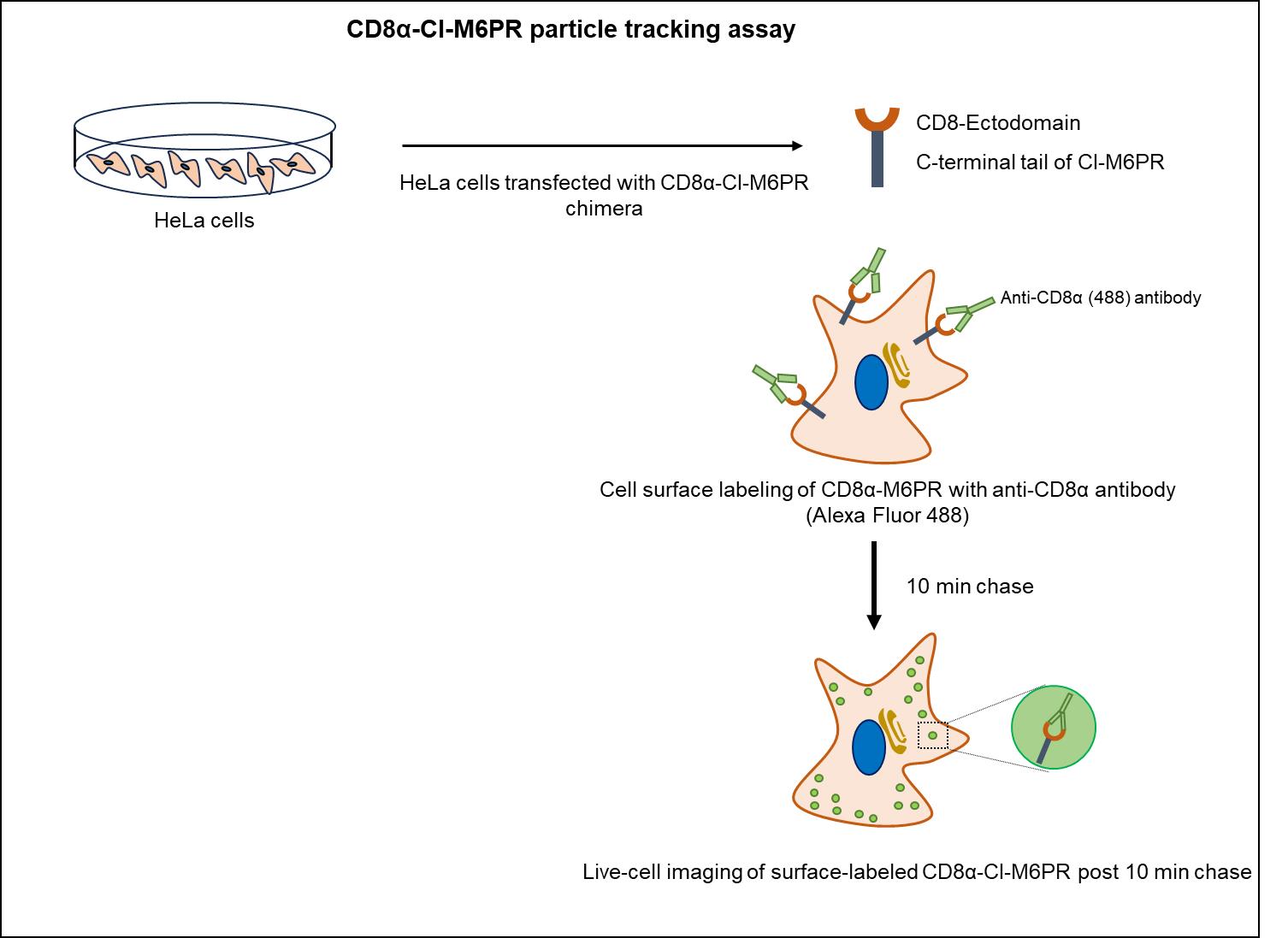
Figure 1. Schematic illustrating the CD8α-CI-M6PR trafficking assay to track motility of CD8α-CI-M6PR endosomes internalized from the cell surface.HeLa cells expressing CD8α-CI-M6PR were incubated with primary (anti-CD8α) and secondary (Alexa Fluor 488-conjugated dye) antibodies on ice to specifically label surface receptors, followed by live-cell imaging in warm media at 37 °C after 10 min of chase.
Materials and reagents
Biological materials
70%–80% confluent HeLa cells (ATCC)
Note: We have used HeLa cells for this analysis because they are easy to transfect and considerably flat for 2D analysis of endosome motility; however, other cell lines can also be used.
Reagents
Plasmid CD8α-CI-M6PR-pIRES Neo2 (a kind gift from Prof. Matthew Seaman)
GibcoTM Dulbecco’s modified Eagle medium (DMEM), high glucose, with GlutaMAXTM, sodium pyruvate (Thermo Fisher Scientific, catalog number: 10569-010), storage: 2–8 °C
Dulbecco’s phosphate buffered saline (DPBS), without calcium and magnesium (Lonza Bioscience, catalog number: 17-512F), storage: 15–30 °C
Opti-MEM® reduced serum media (Thermo Fisher Scientific, GibcoTM, catalog number: 11058021), storage: 2–8 °C
Heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific, Gibco TM, catalog number: 10082147), storage: -20 °C
Antibiotic-antimycotic (Thermo Fisher Scientific, GibcoTM, catalog number: 15240-062), storage: -20 °C
GibcoTM 1 M HEPES (Thermo Fisher Scientific, catalog number: 15630-080), storage: 2–8 °C
GibcoTM 100× MEM non-essential amino acid solution (Thermo Fisher Scientific, catalog number: 11140050)
GibcoTM DMEM, high glucose, HEPES, no phenol red (Thermo Fisher Scientific, catalog number: 21063029), storage: 2–8 °C
X-tremeGENETM HD transfection reagent (Roche® Life Science Products, catalog number: 6366236001)
siRNA transfection reagent Dharmafect-1 (GE Healthcare, catalog number: T-2001-03)
Mouse anti-human CD8 monoclonal antibody (BD PharmingenTM, catalog number: 555631), storage: 4 °C
Alexa Fluor 488-conjugated goat anti-mouse IgG (Thermo Fisher Scientific, catalog number: A11029), storage: 4 °C
Citrate anhydrous (HiMedia, catalog number: GRM-1023)
Sodium citrate tribasic dihydrate (Tri sodium citrate dihydrate) (Sigma, catalog number: S4641)
Citric acid buffer, pH 4.5 (see Recipes)
Recipes
Citric acid buffer, pH 4.5
Reagent Volume Citric acid anhydrous 100 mL Tri-sodium citrate dihydrate 100 mL Total 200 mL 0.1 M Citric acid anhydrous
Reagent Quantity Citric acid anhydrous 2.626 g Total 125 mL 0.1 M Tri-Sodium Citrate dihydrate
Reagent Quantity Tri-sodium citrate dihydrate 3.676 g Total 125 mL Adjust pH to 4.5 with sodium citrate.
Complete medium
Dulbecco’s modified Eagle medium (DMEM), high glucose, with GlutaMAXTM, sodium pyruvate supplemented with 10% FBS, 10 mM HEPES, 1× antibiotic-antimycotic, and 1× MEM non-essential amino acid solution
Laboratory supplies
35 mm μ-dish, high glass-bottom live-imaging dishes (ibidi, catalog number: 81158)
BD Falcon 15 mL centrifuge tubes (Corning, Falcon®, catalog number: 352096)
BD Falcon 35 mm cell culture dish (Corning, Falcon®, catalog number: 353001)
Equipment
Cell culture incubator (Eppendorf, New BrunswickTM, model: Galaxy ® 170 R)
Confocal microscope (ZEISS, model: LSM 710)
HulaMixerTM sample mixer (Thermo Fisher Scientific, catalog number: 15920D)
Software and datasets
Fiji software ( https://imagej.net/software/fiji/) 2.15.1
ZEN software, ZEN 2012 v. 8.0.1.273 (ZEISS)
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Rawat, S. and Sharma, M. (2024). CD8α-CI-M6PR Particle Motility Assay to Study the Retrograde Motion of CI-M6PR Receptors in Cultured Living Cells. Bio-protocol 14(9): e4979. DOI: 10.21769/BioProtoc.4979.
- Rawat, S., Chatterjee, D., Marwaha, R., Charak, G., Kumar, G., Shaw, S., Khatter, D., Sharma, S., de Heus, C., Liv, N., et al. (2023). RUFY1 binds Arl8b and mediates endosome-to-TGN CI-M6PR retrieval for cargo sorting to lysosomes. J. Cell Biol. 222(1). https://doi.org/10.1083/jcb.202108001
分类
细胞生物学 > 细胞成像 > 活细胞成像
细胞生物学 > 基于细胞的分析方法 > 转运
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













