- EN - English
- CN - 中文
Genetic Knock-Ins of Endogenous Fluorescent Tags in RAW 264.7 Murine Macrophages Using CRISPR/Cas9 Genome Editing
利用CRISPR/Cas9基因组编辑技术在RAW 264.7小鼠巨噬细胞中敲入内源性荧光标签
(§Technical contact: bnaigles@ucsd.edu) 发布: 2024年03月20日第14卷第6期 DOI: 10.21769/BioProtoc.4960 浏览次数: 4254
评审: David PaulManoj B. MenonAnonymous reviewer(s)

相关实验方案

利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1253 阅读
Abstract
CRISPR/Cas9 genome editing is a widely used tool for creating genetic knock-ins, which allow for endogenous tagging of genes. This is in contrast with random insertion using viral vectors, where expression of the inserted transgene changes the total copy number of a gene in a cell and does not reflect the endogenous chromatin environment or any trans-acting regulation experienced at a locus. There are very few protocols for endogenous fluorescent tagging in macrophages. Here, we describe a protocol to design and test CRISPR guide RNAs and donor plasmids, to transfect them into RAW 264.7 mouse macrophage-like cells using the Neon transfection system and to grow up clonal populations of cells containing the endogenous knock-in at various loci. We have used this protocol to create endogenous fluorescent knock-ins in at least six loci, including both endogenously tagging genes and inserting transgenes in the Rosa26 and Tigre safe harbor loci. This protocol uses circular plasmid DNA as the donor template and delivers the sgRNA and Cas9 as an all-in-one expression plasmid. We designed this protocol for fluorescent protein knock-ins; it is best used when positive clones can be identified by fluorescence. However, it may be possible to adapt the protocol for non-fluorescent knock-ins. This protocol allows for the fairly straightforward creation of clonal populations of macrophages with tags at the endogenous loci of genes. We also describe how to set up imaging experiments in 24-well plates to track fluorescence in the edited cells over time.
Key features
• CRISPR knock-in of fluorescent proteins in RAW 264.7 mouse macrophages at diverse genomic loci.
• This protocol is optimized for the use of the Neon transfection system.
• Includes instructions for growing up edited clonal populations from single cells with one single-cell sorting step and efficient growth in conditioned media after cell sorting.
• Designed for knocking in fluorescent proteins and screening transfected cells byFACS, but modification for non-fluorescent knock-ins may be possible.
Graphical overview
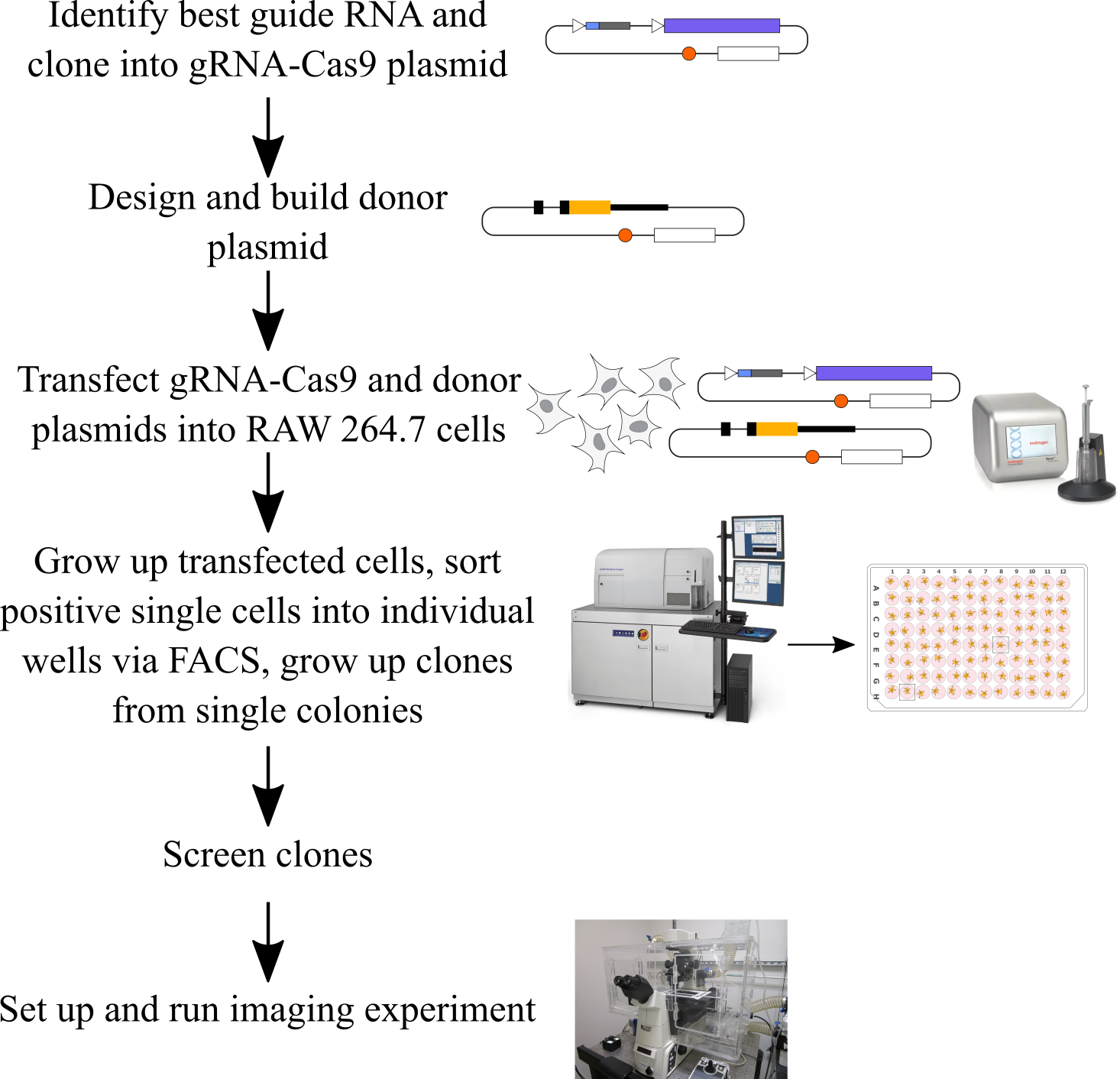
Background
The use of CRISPR/Cas9 genome editing to insert DNA into the genome at a specific locus using the cell’s endogenous homology-directed repair (HDR) pathway is a valuable approach for modifying the genome of a cell [1,2]. One specific use of this technology is to endogenously tag genes with fluorescent proteins, enabling the study of gene expression and protein localization in live single cells. In contrast to the use of viral vectors to randomly insert transgenes into the genome, endogenous tagging ensures that the tagged protein is expressed using all endogenous regulation, including any trans-acting regulatory factors, which is useful for studying the mechanisms of gene expression. Endogenous tagging also ensures that the copy number of the protein remains the same in tagged and untagged cells, in contrast to the overexpression that occurs when using transgenes. There are existing knock-in protocols for many common easy-to-transfect cells lines, such as human RPE1, HCT116 [3], HeLa, and U2-OS cells [4], human iPSCs [5], and mouse embryonic stem cells [6]. In macrophages, there are existing protocols for knock-out CRISPR screens in mouse bone marrow–derived macrophages (BMDMs) [7] and in mouse RAW 264.7 cells [8], as well as for targeted knock-outs using Cas9-ribonucleoprotein complex nucleofection in mouse BMDMs [9]. A recent study reported a protocol for inserting transgenes into the Rosa26 locus of RAW 264.7 cells using CRISPR/Cas9 technology coupled to electroporation of plasmid DNA and growing up a bulk population of edited cells [10]. We developed the protocol that we report here to use CRISPR/Cas9 genome editing to create clonal populations of macrophages with knock-in fluorescent tags at diverse genomic loci in order to study stimulus-responsive gene expression at a single-cell level by imaging the fluorescent protein over time in single cells [11]. We base our CRISPR design on the strategy first developed by Ran and colleagues and use their pSpCas9 plasmid and one-step restriction-ligation cloning strategy for the guide RNA (gRNA) [12]. In this protocol, we use the Neon transfection system to deliver the gRNA, Cas9, and HDR donor sequences as circular plasmid DNA, coupled to a simple strategy for growing up clonal populations following a single single-cell sorting step. In this paper, we use a knock-in of YFP at the C-terminus of the IRF1 protein as our example, but we have used this approach for at least six different loci. This protocol works best for fluorescent proteins because FACS is used to sort positive cells, and positive cells are quite rare. However, it may be possible to adapt this protocol to non-fluorescent knock-ins in the future. At the end of this protocol, we also describe how we prepare an imaging experiment with the edited cells.
Materials and reagents
Biological materials
RAW 264.7 mouse macrophage-like cells (ATCC, catalog number: TIB-71)
NIH3T3 mouse embryonic fibroblasts (ATCC, catalog number: CRL-1658)
pSp-Cas9(BB)-2A-Puro (PX459) plasmid (Addgene, catalog number: 62988)
pUC19 plasmid (Addgene, catalog number: 50005)
Neon Transfection System 100 μL kit (Thermo Fisher, catalog number: MPK10025)
HyCloneTM classical liquid media, Dulbecco’s modified Eagles medium, high glucose, GE healthcare cell culture (DMEM)/high: with 4500 mg/L glucose and 4.0 mM L-Glutamine, without sodium pyruvate (VWR, catalog number: 16750-072)
Fetal bovine serum (FBS) (Fisher, catalog number: MT35010CV)
DPBS, no calcium, no magnesium (Thermo Fisher, catalog number: 14190250)
Penicillin-Streptomycin solution, 100×, 10,000 IU penicillin, 10,000 μg/mL streptomycin (Fisher, catalog number: MT30002CI)
Gibco DMEM, high glucose, no glutamine, no phenol red (Thermo Fisher, catalog number: 31053036)
Gibco L-Glutamine (200 mM) (Thermo Fisher, catalog number: 25030081)
T4 DNA ligase reaction buffer (NEB, catalog number: B0202S)
T4 polynucleotide kinase (PNK) (NEB, catalog number: M0201S)
10× Tango buffer (Thermo Fisher, catalog number: BY5)
Dithiothreitol (DTT) (Millipore Sigma, catalog number: D9799-5g)
10 mM ATP (NEB, catalog number: P0756S)
Bbs1-HF (NEB, catalog number: R3539S)
T7 DNA ligase (enzymatics, catalog number: L6020L)
Autoclaved MilliQ water
NEB 5-alpha competent E. coli (high efficiency) (NEB, catalog number: C2987I)
Opti-MEM reduced serum medium (Thermo Fisher, catalog number: 31985070)
Lipofectamine 2000 transfection reagent (Thermo Fisher, catalog number: 11668027)
Puromycin dihydrochloride (Millipore Sigma, catalog number: P8833-25MG)
Macherey-Nagel Nucleobond Xtra Midi Plus EF kit (Macherey Nagel, catalog number: 740422.5)
QIAprep Spin Miniprep kit (Qiagen, catalog number: 27104)
Zymo Quick-gDNA MiniPrep kit capped columns (Zymo Research, catalog number: D3024)
QIAquick Gel Extraction kit (Qiagen, catalog number: 28704)
Zymo DNA Clean & Concentrator-5 kit capped columns (Zymo Research, catalog number: D4013)
Phusion High-Fidelity DNA Polymerase-100u (NEB, catalog number: M0530S)
NEBuilder HiFi DNA Assembly Master Mix (NEB, catalog number: E2621S)
EDTA powder (Sigma, catalog number: E9884)
HEPES 1 M (Gibco, catalog number: 15630080)
Luria Broth (LB)-carbenicillin agar plates, with carbenicillin at 100 μg/mL
Luria Broth (LB)
Carbenicillin (Fisher BioReagents, catalog number: BP26485), stock at 100 mg/mL
Corning 0.25% Trypsin, 0.1% EDTA in HBSS w/o calcium, magnesium, and sodium bicarbonate; 6/PK 25-053-CI (Fisher, catalog number: MT25053CI)
Complete DMEM (See Recipes)
Conditioned DMEM (See Recipes)
Antibiotic-free DMEM (See Recipes)
0.1 M EDTA solution (See Recipes)
FACS sorting buffer (See Recipes)
Phenol Red–free DMEM (See Recipes)
1,000× Puromycin solution (See Recipes)
Recipes
Complete DMEM
Reagent Final concentration Quantity or Volume HyCloneTM DMEM N/A 445 mL FBS 10% 50 mL Penicillin-Streptomycin solution, 100× 1% 5 mL Total n/a 500 mL Combine all ingredients and pass through a 0.2 μm filter bottle (Corning).
Conditioned DMEM
Reagent Final concentration Quantity or Volume Supernatant from RAW 264.7 culture (RAW 264.7 cells split from a confluent plate at 1:8 density in complete DMEM with 10% FBS, 1% pen/strep, supernatant is collected after 24 h) 50% 100 mL HyCloneTM DMEM n/a 69 mL FBS 20% 30 mL Penicillin-Streptomycin solution, 100x 1% 1 mL Total n/a 200 mL Combine all ingredients and pass through a 0.2 μm filter bottle (Corning).
Antibiotic-free DMEM
Reagent Final concentration Quantity or Volume HyCloneTM DMEM n/a 450 mL FBS 10% 50 mL Total n/a 500 mL Combine all ingredients and pass through a 0.2 μm filter bottle (Corning).
0.1 M EDTA
Reagent Final concentration Quantity or Volume Autoclaved MilliQ water n/a Make up to 50 mL EDTA powder 0.1 M 1.46 g Total n/a 50 mL To make the 0.1 M EDTA, dissolve the EDTA powder in 45 mL of autoclaved MilliQ water, then top off to 50 mL once totally dissolved for a total volume of 50 mL. Filter this solution through a 0.2 μm syringe filter (Acrodisc) before use.
FACS sorting buffer
Reagent Final concentration Quantity or Volume DPBS n/a 47.75 mL EDTA 0.1 M (from Recipe 4) 1 mM 500 µL HEPES 1 M 25 mM 1.25 mL FBS 1 % 500 µL Total n/a 50 mL To make the FACS sorting buffer, combine all reagents and then pass through a 0.2 μm syringe filter (Acrodisc).
Note: Other compositions of FACS sorting buffer will likely also work.
Phenol Red–free DMEM
Reagent Final concentration Quantity or Volume Gibco DMEM, no phenol red N/A 440 mL FBS 10% 50 mL Penicillin-Streptomycin solution, 100× 1% 5 mL L-Glutamine (200 mM) 2 mM 5 mL Total n/a 500 mL Combine all ingredients and pass through a 0.2 μm filter bottle (Corning).
1,000× Puromycin solution (1 mg/mL)
Reagent Final concentration Quantity or Volume Autoclaved MilliQ water n/a Make up to 10 mL Puromycin dihydrochloride 1 mg/mL 10 mg Total n/a 10 mL To make the 1,000× puromycin, dissolve the puromycin dihydrochloride in 9 mL of autoclaved MilliQ water, then top off to 10 mL once totally dissolved for a total volume of 10 mL. Filter this solution through a 0.2 μm syringe filter (Acrodisc) before use and store at -20 °C in 1 mL aliquots.
Laboratory supplies
Falcon 5 mL round bottom polystyrene test tube, with cell strainer snap cap (Falcon, catalog number: 352235)
GenClone 96-well cell culture plates flat bottom wells, TC treated (Genesee, catalog number: 25-109)
Falcon tissue culture plates 24 well (Fisher, catalog number: 087721H)
GenClone 6-well TC treated plates (Genesee, catalog number: 25-105MP)
GenClone TC treated dishes, 100 × 20 mm vented (Genesee, catalog number: 25-202)
Acrodisc syringe filter 0.2 µm Supor membrane, low protein binding, non-pyrogenic, PN 4612 (VWR, catalog number: 28143-310)
Pipette tips, serological pipettes, Eppendorf tubes, PCR tubes, 50 mL conical tubes, 15 mL conical tubes, 50 mL syringes
Cell scraper (e.g., VWR, catalog number: 75799-934)
Corning® 500 mL vacuum filter/storage bottle system, 0.22 µm pore 33.2 cm PES membrane, sterile (Corning, catalog number: 431097)
Equipment
Neon transfection system (Thermo Fisher, model: MPK5000)
PCR thermocycler (e.g., Thermo Fisher, model: ProFlex PCR System Thermocycler)
Hemacytometer (e.g., Hausser Scientific Hemacytometer Thermo Fisher, model: S17040)
Agarose gel electrophoresis rigs
Fluorescence microscope (e.g., Nikon, model: Eclipse Ti)
FACS machine (e.g., BD, model: FACSAria Fusion)
Benchtop mini centrifuge (e.g., MyFuge, model: Southern Labware C1012)
Benchtop microcentrifuge (e.g., Eppendorf, model: 5420)
Software and datasets
ApE (plasmid editor) v3.1.3 November 11, 2022. Free, but you can equally well use a paid cloning software such as SnapGene if you prefer
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Naigles, B., Soroczynski, J. and Hao, N. (2024). Genetic Knock-Ins of Endogenous Fluorescent Tags in RAW 264.7 Murine Macrophages Using CRISPR/Cas9 Genome Editing. Bio-protocol 14(6): e4960. DOI: 10.21769/BioProtoc.4960.
- Naigles, B., Narla, A. V., Soroczynski, J., Tsimring, L. S. and Hao, N. (2023). Quantifying dynamic pro-inflammatory gene expression and heterogeneity in single macrophage cells. J. Biol. Chem. 299(10), 105230. https://doi.org/10.1016/j.jbc.2023.105230
分类
免疫学 > 免疫细胞成像 > 荧光显微技术
细胞生物学 > 细胞工程 > CRISPR-cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










