- EN - English
- CN - 中文
Dissecting the Mechanical Control of Mitotic Entry Using a Cell Confinement Setup
利用单细胞限制器解析有丝分裂进入的机械控制
发布: 2024年03月20日第14卷第6期 DOI: 10.21769/BioProtoc.4959 浏览次数: 2727
评审: Rajesh RanjanSevgi OnalAnkur GargAnonymous reviewer(s)
Abstract
Proliferating cells need to cope with extensive cytoskeletal and nuclear remodeling as they prepare to divide. These events are tightly regulated by the nuclear translocation of the cyclin B1-CDK1 complex, that is partly dependent on nuclear tension. Standard experimental approaches do not allow the manipulation of forces acting on cells in a time-resolved manner. Here, we describe a protocol that enables dynamic mechanical manipulation of single cells with high spatial and temporal resolution and its application in the context of cell division. In addition, we also outline a method for the manipulation of substrate stiffness using polyacrylamide hydrogels. Finally, we describe a static cell confinement setup, which can be used to study the impact of prolonged mechanical stimulation in populations of cells.
Key features
• Protocol for microfabrication of confinement devices.
• Single-cell dynamic confinement coupled with high-resolution microscopy.
• Static cell confinement protocol that can be combined with super-resolution STED microscopy.
• Analysis of the mechanical control of mitotic entry in a time-resolved manner.
Graphical overview
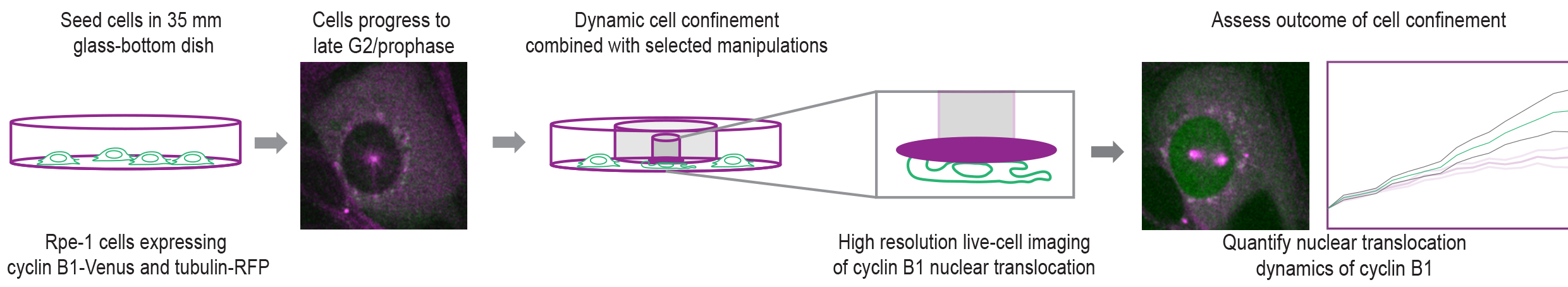
Background
As proliferating cells prepare to enter mitosis, they undergo a series of biochemical and morphological changes that contribute to the assembly of the mitotic machinery and the efficiency of chromosome segregation [1]. Many of these changes are regulated by the activity and subcellular localization of the cyclin B1-CDK1 complex [2,3]. Notably, nuclear translocation of cyclin B1 seems to be essential to ensure full activity of the complex, as it stimulates its own nuclear import [3] through the nuclear pore complexes, triggering a positive feedback mechanism [4]. While the biochemical pathways that regulate the activity and localization of the cyclin B1-CDK1 complex have been well established [5], the contribution of mechanical forces to these processes remained unknown. We have recently identified a tension-dependent signal on the prophase nucleus that fine-tunes cyclin B1 translocation across the nuclear pores, setting the time for mitotic entry [6]. We further show that this signal relies on actomyosin contractility that leads to an unfolding of the nucleus and increased tension on the nuclear envelope (NE). Overall, we propose that mechanical forces are an important contributor for mitotic entry.
Here, we describe a protocol that enables the mechanical manipulation of cells during mitotic entry with high temporal and spatial resolution in single cells undergoing mitosis. This method allows the implementation of cell confinement techniques [7] in combination with high- (spinning-disk) and super-resolution (STED) microscopy to probe how mitotic entry is mechanically regulated. We provide step-by-step instructions on cell seeding, preparation of the confinement slides, assembly of the confinement devices, live-cell imaging, and data analysis. Moreover, we explain how to generate coverslips coated with polyacrylamide (PAA) hydrogels of calibrated stiffness that can be used in combination with high-resolution spinning-disk live-cell imaging. Finally, we describe an experimental setup that allows the long-term confinement of cell populations for fixed-cell immunofluorescence analysis. While these are used in the context of cell division, these methods could be easily adapted to other experimental settings.
Materials and reagents
Biological materials
RPE-1 parental cell line, already available in the lab
RPE-1 expressing endogenously tagged with H2B-GFP and tubulin mRFP, already available in the lab
RPE-1 cell line expressing exogenous cyclinB1-Venus (gift from Jonathon Pines)
RPE-1 cell line expressing cyclinB1-Venus/tubulin-mRFP, generated in our lab by transduction with lentiviral vectors containing pRRL-mRFP-α-tubulin
shRNAi-ROCK1-LV to deplete ROCK1 (gift from João Relvas)
RPE-1 cell line expressing exogenous cGAS-GFP (gift from Matthieu Piel)
RPE-1 cell line expressing GFP-NLS/tubulin-mRFP, created by lentiviral transduction using a pCDJ-NLS-copGFP-EF1-BlastiS plasmid (Addgene, catalog number: 132772)
RPE-1 cell lines expressing exogenous LBR-mCherry and endogenous cyclinB1-Venus/LBR-mCherry, generated by lentiviral transfection using a plasmid pWPT LBR-mCherry (gift from Stephen Royle)
DN-KASH plasmid (gift from Edgar Gomes)
Rap1Q63E plasmid (gift from Jean de Gunzburg)
Reagents
RO-3306 (Santa Cruz Biotechnology, catalog number: sc-358700A), used at 9 mM for 16 h
Importazole (gift from Helder Maiato), used at 40 mM for 2 h
Y-37632 (Sigma-Aldrich, catalog number: Y0503), used at 5 mM for 30 min
AACOCF3 (TOCRIS, catalog number: 1462), used at 20 μM for 30 min
BAPTA-AM (Abcam, catalog number: ab120503), used at 10 μM for 15 min
2APB (Abcam, catalog number: ab120124), used at 10 μM for 30 min
Para-nitro-blebbistatin (MotorPharma, catalog number: 1621326-32-6), used at 50 μM for 30 min
ML-7 (Merck, catalog number: I2764), used at 50 μM for 30 min
Cytochalasin D (Biogen Cientifica, catalog number: TO-1233), used at 0.5 μM for 30 min
Nocodazole (Merck, catalog number: M1404), used at 3.3 μM for 30 min
BI2536 (Axon Med Chem, catalog number: AXON 1129), used at 200 mM for 2 h
Etoposide (Selleck Chemicals CO., catalog number: S1225), used at 1 μM for 2 h
Thymidine (Sigma-Aldrich, catalog number: T1895), used at 2 mM for 16 h followed by a 10 h release
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D4540)
Lipofectamine 2000 (Invitrogen, catalog number: 11668019)
Lipofectamine RNAiMAX (Invitrogen, catalog number: 13778150)
Dulbecco’s Modified Eagle Medium (DMEM) (Corning, catalog number: 10-013-CV)
Fetal bovine serum (FBS) (Gibco, catalog number: 11550356)
Fibronectin (FBN) (Sigma, catalog number: F1141)
Leibovitz’s-L15 medium (Life Technologies, catalog number: 11415064)
Antibiotic-Antimycotic 100× (Life Technologies, catalog number: 15240096)
Bind-Silane (Sigma, catalog number: M6514)
Acetic acid (glacial) 100% (Merck, catalog number: 1.00063)
40% (w/v) acrylamide stock solution (Bio-Rad, catalog number: 1610140)
2% (w/v) bis-acrylamide stock solution (Bio-Rad, catalog number: 1610142)
PDMS crosslinker A + B (Momentive, catalog number: RTV615)
Isopropanol (Merck, catalog number: 1.09634)
dH2O
Trypsin (TrypLE Express Enzyme 1×) (Thermo Fisher Scientific, catalog number: 12605010)
Trypan Blue solution 0.4% (Gibco, catalog number: 15250061)
Sodium hydrogen carbonate (NaHCO3) (Sigma-Aldrich, catalog number: S6014)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 31434)
Potassium chloride (KCl) (Merck, catalog number: 1.04936)
Dissodium hydrogen phosphate (Na2HPO4) (Merck, catalog number: 1.06586)
Potassium dihydrogen phosphate (KH2PO4) (Merck, catalog number: 1.04873)
Absolute ethanol (Sigma-Aldrich, catalog number: E7148)
Hydrochloric acid (HCl) 37% (Merck, catalog number: 1.00317)
Sodium hydroxide (NaOH) (Merck, catalog number: 1.06498)
Solutions (see note 1)
10% acetic acid (see Recipes)
Bind-silane solution (see Recipes)
Phosphate buffered saline (PBS) 10× (see note 2) (see Recipes)
Fibronectin (FBN) solution (see note 3) (see Recipes)
70% ethanol (see Recipes)
Recipes
10% acetic acid
Reagent Final concentration Quantity or Volume Acetic acid (glacial) 100% 10% (v/v) 10 mL Absolute ethanol n/a 90 mL Total n/a 100 mL Bind-silane solution
Reagent Final concentration Quantity or Volume Acetic acid 10% 5% 2.5 mL Bind silane 0.3% 15 μL Absolute ethanol n/a 2.485 mL Total n/a 5 mL Phosphate buffered saline (PBS) 10×
Reagent Final concentration Quantity or Volume NaCl 1.37 M 80 g KCl 0.027 M 2 g Na2HPO4 0.1 M 14.4 g KH2PO4 0.018 M 2.4 g H2O n/a Top up volume for 1 L Total n/a 1 L Fibronectin (FBN) solution
Reagent Final concentration Quantity or Volume FBN 25 mg/mL 25 mg NaHCO3 pH 8.6a 100 mM Top up volume for 1 mL Total n/a 1 mL a Dilute 84 mg of NaHCO3 (MW = 84.01 g/mol) in 10 mL of ddH2O and adjust the pH to 8.6. Store at room temperature.
Ethanol 70% solution
Reagent Final concentration Quantity or Volume Absolute ethanol 70% 700 mL dH2O n/a Top up volume for 1 L Total n/a 1 L
Laboratory supplies
FluoroDishes (WPI, catalog number: FD-35-100)
Round glass coverslips 10 mm thickness #1.5 (Fisher Scientific, catalog number: NC1272767)
Square glass coverslips 22 mm × 22 mm thickness #1.5 (Corning, catalog number: 2850-22)
Microfabricated coverslips with confining pillars
6-well cell culture plate (Corning, catalog number: CLS3516)
6-well static confiner (4DCell, catalog number: CSOW 620)
T25 cell culture flasks (Sarstedt, catalog number: 83.3910)
Scotch Magic tape (3M, catalog number: 8-1933)
Parafilm M (Fisher Scientific, catalog number: 10018130)
Scalpel blade or razor blade
Fine forceps (Fine Science Tools, catalog number: 11254-20)
SU-8 mold with micropillar design (either custom-designed or ordered from MesoBioTech, France)
Harris Uni-Core puncher (0.75 mm) (WPI, catalog number: 504529)
Disposable 5 mL sterile pipettes (nerbe plus, catalog number: 12-441-9105)
Disposable 10 mL sterile pipettes (nerbe plus, catalog number: 12-461-9108)
Disposable 50 mL centrifuge tubes (Corning, catalog number: 734-1868)
Disposable 15 mL centrifuge tubes (Corning, catalog number: 734-1812)
Cell counting slides (Bio-Rad, catalog number: 1450011)
Equipment
Temperature-controlled TE2000 microscope equipped with a modified Yokogawa CSU-X1 spinning-disk head (Yokogawa Electric), an electron multiplying iXon+ DU-897 EM-CCD camera (Andor), and a filter wheel (Nikon)
Immersion oil 60× 1.4NA Plan-Apo DIC objective (Nikon)
Dynamic cell confiner (adapted from Le Berre et al. [7])
AF-1 dual vacuum/pressure controller (Elveflow)
6-well confinement device (4DCell, France)
Low-pressure plasma cleaner (Zepto, Diener Electronics, Germany)
Centrifuge
Hot plate
pH meter
Electronic TC10TM/TC20TM cell counter device
Vacuum degasser (optional)
37 °C humidified CO2 (5%) incubator
Laminar flow hood
Software and datasets
NIS Elements AR software (Nikon)
ImageJ (version 1.54)
Custom-designed MATLAB algorithm for nuclear pore analysis (The MathWorks Inc, USA; v2018b)
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Dantas, M., Vareiro, D. and Ferreira, J. G. (2024). Dissecting the Mechanical Control of Mitotic Entry Using a Cell Confinement Setup. Bio-protocol 14(6): e4959. DOI: 10.21769/BioProtoc.4959.
- Dantas, M., Oliveira, A., Aguiar, P., Maiato, H. and Ferreira, J. G. (2022). Nuclear tension controls mitotic entry by regulating cyclin B1 nuclear translocation. J. Cell Biol. 221(12): e202205051.
分类
细胞生物学 > 细胞结构 > 细胞核
细胞生物学 > 单细胞分析
细胞生物学 > 基于细胞的分析方法 > 细胞吞排作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link