- EN - English
- CN - 中文
Preparation and Purification of β-1,3-glucan-Linked Candida glabrata Cell Wall Proteases by Ion-Exchange Chromatography, Gel Filtration, and MDPF-Gelatin-Zymography Assay
通过离子交换色谱、凝胶过滤和MDPF-明胶-酶谱分析法准备和纯化β-1,3-葡聚糖连接的光滑念珠菌细胞壁蛋白酶
(*contributed equally to this work) 发布: 2024年03月20日第14卷第6期 DOI: 10.21769/BioProtoc.4958 浏览次数: 1711
评审: Alba BlesaSuresh PantheeSascha Brunke

相关实验方案

通过制备连续聚丙烯酰胺凝胶电泳和凝胶酶谱分析法纯化来自梭状龋齿螺旋体的天然Dentilisin复合物及其功能分析
Pachiyappan Kamarajan [...] Yvonne L. Kapila
2024年04月05日 2096 阅读
Abstract
Candida glabrata is an opportunistic pathogen that may cause serious infections in an immunocompromised host. C. glabrata cell wall proteases directly interact with host cells and affect yeast virulence and host immune responses. This protocol describes methods to purify β-1,3-glucan-bonded cell wall proteases from C. glabrata. These cell wall proteases are detached from the cell wall glucan network by lyticase treatment, which hydrolyzes β-1,3-glucan bonds specifically without rupturing cells. The cell wall supernatant is further fractioned by centrifugal devices with cut-offs of 10 and 50 kDa, ion-exchange filtration(charge), and gel filtration (size exclusion). The enzymatic activity of C. glabrata proteases is verified with MDPF-gelatin zymography and the degradation of gelatin is visualized by loss of gelatin fluorescence. With this procedure, the enzymatic activities of the fractions are kept intact, differing from methods used in previous studies with trypsin digestion of the yeast cell wall. The protein bands may be eventually located from a parallel silver-stained gel and identified with LC–MS/MS spectrometry. The advantage of this methodology is that it allows further host protein degradation assays; the protocol is also suitable for studying other Candida yeast species.
Key features
• Uses basic materials and laboratory equipment, enabling low-cost studies.
• Facilitates the selection and identification of proteases with certain molecular weights.
• Enables further functional studies with host proteins, such as structural or immune response–related, or enzymes and candidate protease inhibitors(e.g., from natural substances).
• This protocol has been optimized for C. glabrata but may be applied with modifications to other Candida species.
Graphical overview
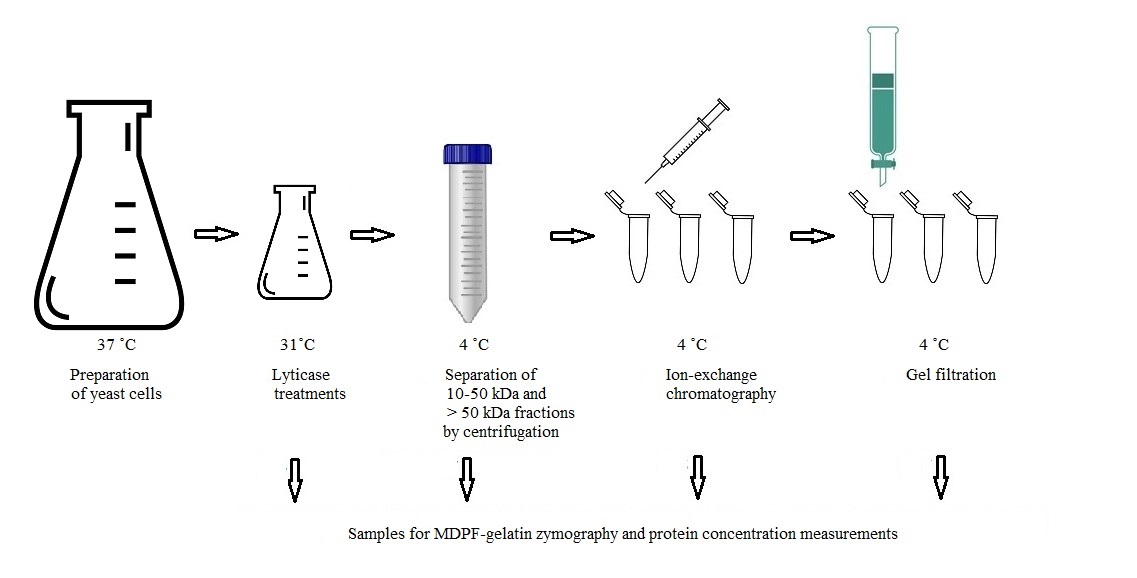
Background
Candida glabrata is an opportunistic pathogen causing infections in vulnerable hosts. As C. glabrata is often resistant to the most used azole antimycotics, the search for novel and safe antimycotics is ongoing. Among the most difficult challenges in understanding yeast pathogenesis is identifying the yeast proteome and virulence factors that affect the course of infection. While the cell wall composition of Candida albicans, the most common opportunistic yeast found in candidiasis, has been quite extensively studied, less is known of C. glabrata’s cell wall proteins, whose key elements and interactions remain undiscovered. The cell wall proteome is the front line when contacting host cells. C. glabrata cell wall structure resembles that of C. albicans and, even more closely, of S. cerevisiae [1–5].
Glycosylphosphatidylinositol (GPI) proteins, covalently bound to the cell wall β-1,6-glucan, as well as proteins linked through a mild-alkali-sensitive linkage to β-1,3-glucan are the two main classes of C. glabrata cell wall proteins, which participate in the interaction with the host [2]. To the last group belong Candida proteins that are delivered to the cell wall from their original intracellular location, named as “moonlighting” proteins. They lack an N-terminal signal sequence guiding formed proteins into the cell wall and are transported there in extracellular vesicles; these have extracellular enzymatic activities in addition to their intracellular housekeeping roles and play multiple functions in host cell interactions. The role of these moonlighting proteins as virulence factors is complex and partially unknown.
The presented protocol describes the enzymatic separation and purification of proteases from C. glabrata cell wall attached to the glucan network (see workflow in Graphical overview). The main objective is to identify novel proteases from the C. glabrata cell wall and enable further analysis of their potential enzymatic activities in host protein degradation. The protocol retains the isolated proteases in their active forms, which is a crucial advantage compared to previous studies in proteomics, allowing the understanding of the functions and interactions with host proteins. The glycosylation of cell wall proteins poses challenges in their identification process: carbohydrates attached to proteins mask them, hampering enzymes used in assays from degrading the protein into a form that may be identified by mass analysis. By detaching the proteases from the cell wall glucan network, it is possible to identify isoenzyme forms by MS/MS. Although dithiothreitol or β-mercaptoethanol have been traditionally used in extracting yeast cell wall proteins, they may cause cell rupture or denaturation of proteins [4]. The use of the β-1,3-glucan specific enzyme lyticase (from Arthrobacter luteus) in cleaving is essential to maintain the proteases in their active form, contrary to conventional non-functional studies. Lyticase has traditionally been used in the degradation of the yeast cell wall, formation of spheroplasts, and transformation procedures, and Candida enzymatic activity may be determined by 7-amino-4-methylcoumarin (AMC)-labeled L-arginine fluorogenic substrate degradation [6].
2-Methoxy-2,4-diphenyl-3(2H)-furanone (MDPF)-gelatin zymography is a convenient method for monitoring the protein purification process. In MDPF-gelatin zymography, the C. glabrata proteases retain their enzymatic activity, because non-denaturing conditions are maintained through the process. The strength of gelatinolysis caused by C. glabrata proteases may be seen as the disappearance of the MDPF-labeled gelatin fluorescence in the SDS-PAGE gel, which may be monitored under UV light. MDPF is a more stable fluorophore compared with FITC or fluorescein [7,8]. MDPF-gelatin zymography is suitable for host protein degradation assays [9]. The lyticase extract is initially separated into coarse fractions with centrifugal devices having cut-off values of 10 and 50 kDa. Further protein charge–based ion-exchange chromatographic separation is optimized regarding pH, and a weak anion-exchange column is used to extract > 50 kDa proteins; further 10–50 kDa and > 50 kDa fractions are obtained by size-exclusion gel filtration. The refining of these fractions enables proper separation, extraction, and identification of the protein bands from silver-stained gels for further mass analyses. The methods used here are easily applied with modifications to study the cell wall proteome in other pathogenic species when searching proteins with specific linkage to the cell wall. In studying the entire proteome of the cell wall, a shotgun approach should be preferred.
Materials and reagents
Biological materials
C. glabrata T-1639, blood isolate (Helsinki University Central Hospital, Helsinki, Finland). Other Candida species may be studied by these methods if there are structural analogies with C. glabrata cell wall protein attachment molecules.
Sabouraud dextrose agar (Lab M, catalog number: LAB009)
Lyticase, 10 kU (Sigma-Aldrich, catalog number: 37340-57-1)
2-Methoxy-2,4-diphenyl-3(2H)-furanone (MDPF) (Sigma-Aldrich, catalog number: 50632-57-0)
Precision Plus ProteinTM standards (Bio-Rad, catalog number: 161-0373)
Tris base (Sigma, catalog number: T-1378)
ZnCl2 (Sigma, catalog number: 208086)
CaCl2 (Sigma, catalog number: 10035-04-8)
Tween 80, BioXtra viscous liquid (Sigma, catalog number: P8074)
BactoTM yeast extract (BD, catalog number: 212750)
Peptone, BactoTM peptone (BD, catalog number: 211677)
Glucose (Merck, catalog number: 108337)
Sodium dodecyl sulphate (SDS) (Fisher Scientific, catalog number: BP166-500)
Sodium azide (NaN3) (Merck, catalog number: 6688), for long-term preservation of ion-exchange columns
Coomassie Brilliant Blue, Serva Blue R (Serva Electrophoresis GmbH, catalog number: 35051)
Sodium tetraborate (STB) (Merck, catalog number: 6308)
Gelatin (Merck, catalog number: 1.04070)
Acetone (Merck, catalog number: 1.00658)
Na2HPO4 (Sigma, catalog number: 137036)
KCl (Sigma, catalog number: PHR1329)
NaCl (Fisher Scientific, catalog number: FLS271500)
KH2PO4 (Merck, catalog number: 104871)
β-mercaptoethanol (Merck, catalog number: M6250)
TEMED (Merck, catalog number: 411019)
Acrylamide 30% (Merck, catalog number: A3574)
EtOH, Ethanolum anhydricum, Aa (Berner, catalog number: 13110124)
HCl 1 M (Riedel-de-Haen, catalog number: 30721)
HCl 37% (Sigma, catalog number: 258148)
NaOH 1 M (Acros Organics, catalog number: 15634890)
Methanol (Fisher, catalog number: M/4057/PB17)
Acetic acid (Fisher, catalog number: A/0400/PB17)
Ammonium persulfate (APS) 10% (Sigma, catalog number: A3678), store in 100 µL aliquots at -20 °C
4x Laemmli Sample Buffer (Bio-Rad, catalog number 1610747)
Glycine (Merck, catalog number: G8898)
Solutions
Yeast extract peptone glucose (YPG) (see Recipes)
MDPF-gelatin (see Recipes)
Separating gel (lower) buffer for zymo gels (see Recipes)
Stacking gel (upper) buffer for zymo gels (see Recipes)
8% MDPF-gelatin SDS-PAGE (see Recipes)
SDS-PAGE running buffer 5× (see Recipes)
Zymo buffer 10× (see Recipes)
Zymo gel incubation buffers (see Recipes)
Coomassie Brilliant Blue (CBB) 0.1% (see Recipes)
IEC start buffer (see Recipes)
IEC elution buffer I (see Recipes)
IEC elution buffer II (see Recipes)
Phosphate-buffered saline (PBS) (see Recipes)
10% SDS (see Recipes)
Lyticase stock (see Recipes)
Recipes
Perform pH adjustments with 1 M NaOH or 1 M HCl. Prepare zymogels as needed and solutions according to Recipes beforehand. Preparing double amounts of Recipes 10–13 is advised.
Yeast extract peptone glucose (YPG)
0.5% yeast extract
1% peptone
1% glucose
Add Milli-Q water (MQ) up to 4 L
Sterilize by autoclave at 121 °C for 15 min
MDPF-gelatin
20 mM STB solution: 0.3814 g of STB per 50 mL of MQ, pH 9.2.
Add 500 mg of gelatin into STB in warm water bath (approximately 37 °C) with magnetic stirring.
Dissolve 25 mg of MDPF into -20 °C acetone on ice and add MDPF into gelatin-STB at room temperature (RT) with magnetic stirring for 1 h protected from light. Divide into 500 µL aliquots and store at -20 °C protected from light.
Separating gel (lower) buffer for zymo gels
800 mL of MQ
187 g of Tris base
Adjust pH to 8.8
4 g of SDS
Add MQ up to 1 L
Stacking gel (upper) buffer for zymo gels
800 mL of MQ
60.5 g of Tris base
Adjust pH to 6.8
4 g of SDS
Add MQ up to 1 L
8% MDPF (1 mg/mL)-gelatin SDS-PAGE (4 gels)
Prepare gels in a fume hood:
Bottom: separating gel
Separating buffer 5 mL (Recipe 3)
30% Acrylamide 5.3 mL
MQ 7.3 mL
MDPF-gelatin (1 mg/mL) 2 mL (Recipe 2)
10% SDS 200 µL
TEMED 20 µL
10% APS 200 µL
Top: stacking gel
Stacking gel buffer 2.5 mL (Recipe 4)
30% Acrylamide 1.35 mL
MQ 5.95 mL
10% SDS 100 µL
TEMED 10 µL
10% APS 100 µL
SDS-PAGE running buffer 5×
Add 4 L of MQ into a Duran bottle
723 g of glycine
150 g of Tris base
20 g of SDS
Adjust pH to 8.3
Add MQ up to 5 L
Zymo buffer 10×
Add 800 mL of MQ into a Duran bottle
60.55 g of Tris base
2 g of NaN3
33 mL of 37% HCl
Adjust pH to 7.5
Add MQ up to 1 L
Zymo gel incubation buffers for 4 gels
Zymo buffer I
360 mL of 1× zymo buffer (dilute from 10× zymo buffer with MQ)
40 mL of 25% Tween 80
Zymo buffer II
270 mL of 1× zymo buffer
30 mL of 25% Tween 80
30 µL of 10 mM ZnCl2
1530 µL of 100 mM CaCl2
Zymo buffer III
300 mL of 1× zymo buffer
30 µL of 10 mM ZnCl2
1530 µL of 100 mM CaCl2
Coomassie Brilliant Blue (CBB), 0.1% (staining and destaining in fume hood)
1 g of Coomassie Brilliant Blue, Serva Blue R
600 mL of MQ
300 mL of methanol
100 mL of acetic acid
Destaining: 40% MeOH + 10% acetic acid in MQ
IEC start buffer (20 mM Tris HCl, pH 8.2)
2.4 g of Tris base
Add 800 mL of MQ
Adjust pH to 8.2
Add MQ up to 1 L
Sterile filter and degas
IEC elution buffer I (20 mM Tris HCl-0.5 M NaCl, pH 8.2)
29.22 g of NaCl
2.4 g of Tris base
Add 800 mL of MQ
Adjust pH to 8.2
Add MQ up to 1 L
Sterile filter and degas
IEC elution buffer II (20 mM Tris HCl- 1 M NaCl, pH 8.2)
14.61 g of NaCl
0.6 g of Tris base
Add 200 mL MQ
Adjust pH to 8.2
Add MQ up to 250 mL
Sterile filter and degas
Phosphate-buffered saline (PBS), pH 7.4
800 mL of MQ
8.0 g of NaCl
1.44 g of Na2HPO4
0.2 g of KCl
0.24 g of KH2PO4
Adjust pH to 7.4 using HCl
Add MQ up to 1 L
Filter and sterilize by autoclavation
10% SDS
80 mL of MQ
10 g of SDS
Add MQ up to 100 mL
Lyticase stock
1.6 mL of MQ
Add 10 kU lyticase
Store at -20 °C in 400 µL aliquots
Laboratory supplies
Sterile plastic inoculation loops (Greiner Bio-One, catalog number: 731171)
Millipore ExpressTM Plus or Filtration unit Stericup® with MILLIPORE Express® PLUS (PES), 0.22 µm (Merck Millipore, catalog number: X342.1, optional method: centrifugation)
Millipore Millex, 0.22 µm (Merck, catalog number: 051581)
Agar plates, plastic Petri dishes, PS (Sigma, catalog number: P5731)
Glass chromatography column (9.5 mL) with stopcock and lower silicon tubing (or Econo Alpha column, Bio-Rad, catalog number: 12009430) and column stand
Glass Erlenmeyers, 2× 3 L, 2× 200 mL
Amicon® Ultra-15 Centrifugal Filter Devices, 10 K and 50 K (Merck Millipore, catalog numbers: UFC901024, UFC905024)
Glass decanters 500 mL
EppendorfTM Snap-Cap Microcentrifuge Safe-LockTM Tubes [Fisher, catalog number: 05-402-18 (1.5 mL) and 05-402-18 (0.5 mL)]
FalconTM tubes, 15 mL, 50 mL (Fisher, catalog numbers: 14-959-53A, 14-432-22)
Micropipettes and sterile pipette tips, 10 µL–5 mL
Aluminum foil
Tube racks for FalconTM and EppendorfTM tubes
HiTrap® IEX Selection Kit (Merck, GE17-6002-33)
Sephadex G-100 and G-200 (Pharmacia Fine Chemicals, catalog numbers: 01-900-1-1861-03, 01-900-1-1863-03) 1 g/40 mL of buffer
10 mL sterile plastic syringes (Fisher, catalog number: 14955459)
Ice, 2 × 5 L plastic dish
Duran bottles, 1 L, 5 L
Funnel that fits gel filtration column
Equipment
Heating cabinet, 31 °C, 37 °C, optional: Melag Incubat® 85 (or Merck My Temp Mini Digital Incubator, catalog number: Z763357) or SureTemp® Dual Convection Incubator (Merck, catalog number: Z742696)
Nanodrop 1000 (or NanoDrop Lite Plus Spectrophotometer, catalog number: NDLPLUSGL)
pH meter (VWR® phenomenal® MU 6100 H, catalog number: 665-0311)
PAGE-running device, 20 µL well volume, 4–6 combs of 10 wells (Bio-Rad, model: Mini-Protean II, catalog number: 165-2940)
Plastic containers (500 mL) for zymo buffer incubation, disposable PP, flat bottom (Marjukka, catalog number: 6410416249018)
Light box for gel visualization, LX104, Lammex (or X-ray viewer; Quirumed, catalog number: 482-6001.S)
Gel scanner, GS-700 densitometer (Bio-Rad, catalog number: 170-7601 with QUANTITY ONE PROGRAM)
Light microscope, 10–40× magnification
Vacuum suction system for degassing buffers and Sephadex gel
UV light for gel visualization and photography (Uvidoc, model: M03 0103 or GelDoc Go Gel Imaging System with Image Lab Touch Software, catalog number: 12009077)
Level mixer (Heidolph Vibramax 100, catalog number: 544-21200-00)
Centrifuge SL16R and adapters for 15 mL tubes (Thermo Scientific, catalog number: 75004030)
Autoclave (Systec, model: D45)
Fume hood (Visukaluste Ltd, model: VISU ProFocus EN 14175-2)
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Pärnänen, P., Sorsa, T., Tervahartiala, T. and Nikula-Ijäs, P. (2024). Preparation and Purification of β-1,3-glucan-Linked Candida glabrata Cell Wall Proteases by Ion-Exchange Chromatography, Gel Filtration, and MDPF-Gelatin-Zymography Assay. Bio-protocol 14(6): e4958. DOI: 10.21769/BioProtoc.4958.
分类
微生物学 > 微生物生物化学 > 蛋白质 > 分离和纯化
生物化学 > 蛋白质 > 分离和纯化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










