- EN - English
- CN - 中文
Efficient Genetic Transformation and Suicide Plasmid-mediated Genome Editing System for Non-model Microorganism Erwinia persicina
非模式微生物桃色欧文氏菌的高效遗传转化及自杀质粒介导的基因组编辑系统
(§ Technical contact) 发布: 2024年03月20日第14卷第6期 DOI: 10.21769/BioProtoc.4956 浏览次数: 2404
评审: Samik BhattacharyaAnonymous reviewer(s)

相关实验方案
I-PREFR:基于反向PCR的无酶单向策略,利用自杀载体在细菌中快速实现无标记染色体基因缺失与重建
Rekha Rana [...] Prabhu B. Patil
2025年05月20日 2318 阅读
Abstract
Erwinia persicina is a gram-negative bacterium that causes diseases in plants. Recently, E. persicina BST187 was shown to exhibit broad-spectrum antibacterial activity due to its inhibitory effects on bacterial acetyl-CoA carboxylase, demonstrating promising potential as a biological control agent. However, the lack of suitable genetic manipulation techniques limits its exploitation and industrial application. Here, we developed an efficient transformation system for E. persicina. Using pET28a as the starting vector, the expression cassette of the red fluorescent protein–encoding gene with the strong promoter J23119 was constructed and transformed into BST187 competent cells to verify the overexpression system. Moreover, suicide plasmid–mediated genome editing systems was developed, and lacZ was knocked out of BST187 genome by parental conjugation transfer using the recombinant suicide vector pKNOCK-sacB-km-lacZ. Therefore, both the transformation and suicide plasmid–mediated genome editing system will greatly facilitate genetic manipulations in E. persicina and promote its development and application.
Key features
• Our studies establish a genetic manipulation system for Erwinia persicina, providing a versatile tool for studying the gene function of non-model microorganisms.
• Requires approximately 6–10 days to complete modification of a chromosome locus.
Graphical overview
Background
Erwinia persicina belongs to the phylum Proteobacteria, class Gammaproteobacteria, order Enterobacteriales, family Enterobacteriaceae, and genus Erwinia [1]. E. persicina has been identified as a soft rot pathogen that causes pink-pigmented soft rot on plant hosts including garlic, onions, barley, Cucurbita pepo, celery, and alfalfa [2–8]. However, E. persicina has exhibited great potential as a biological control agent to inhibit Salmonella enterica contamination in the sprout food industry [9], Fusarium head blight (FHB) severity in wheat [10], and a variety of soil-borne pathogens (Alternaria solani, Sclerotinia sclerotiorum, Rhizoctonia solani, and Fusarium proliferatum) in potato dextrose agar plates [11]. Recently, E. persicina BST187 was isolated from the rhizosphere of tomato and exhibited broad-spectrum antibacterial activity due to its inhibitory effects on bacterial acetyl-CoA carboxylase [12].
The construction and application of chassis modification of E. persicina has not yet been studied, except by Zhao et al. [13]. Efficient genetic transformation and genome editing systems will be beneficial for exploring the pathogenic mechanisms of E. persicina to provide a fundamental basis for control strategies. Moreover, tools developed for the genetic manipulation of E. persicina will provide excellent cell factories for diverse industrial applications such as andrimids.
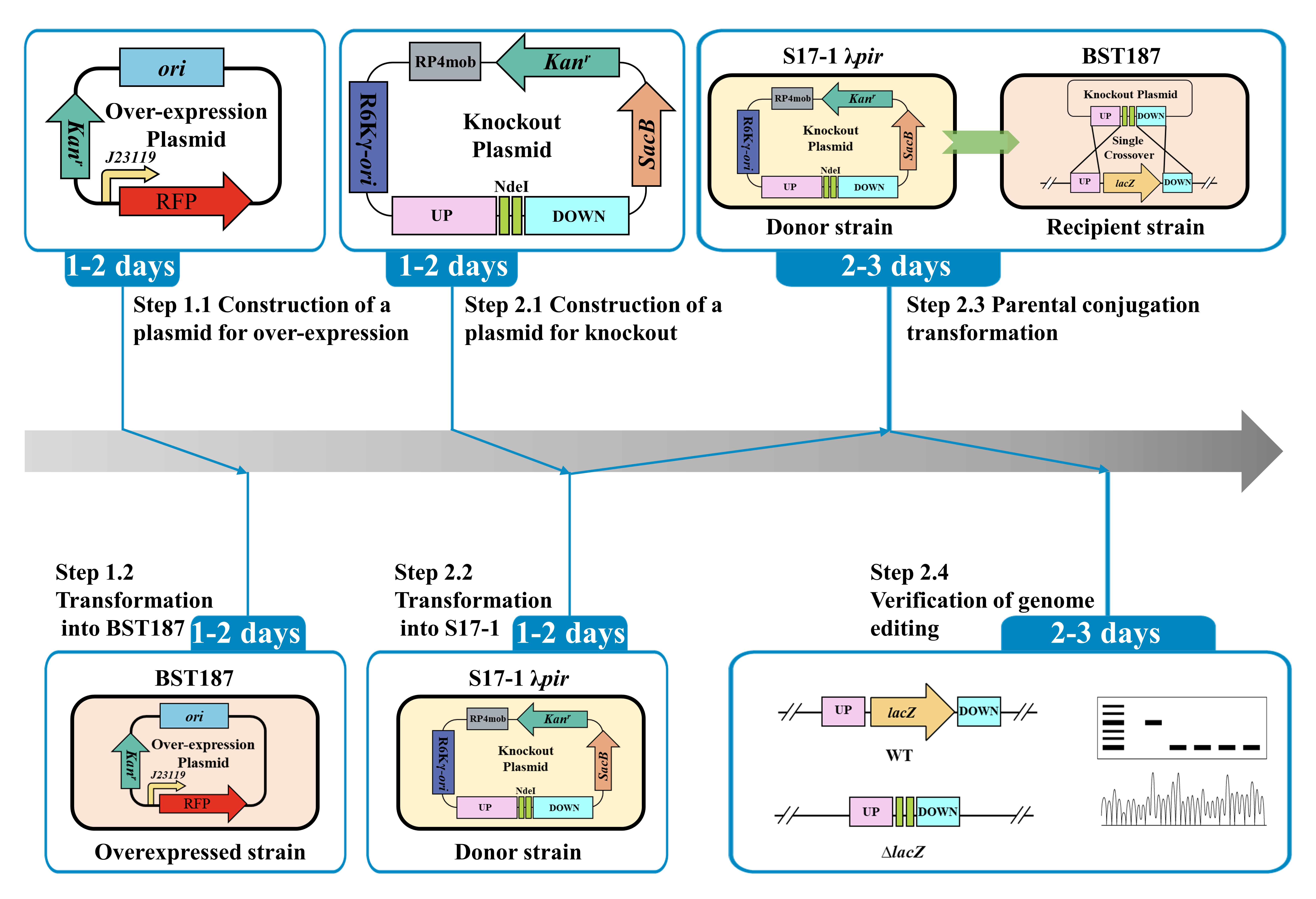
In this study, the details of the BST187 chassis modification system are described. We initially identify the design flaws with the pET series of expression plasmids used for BST187, as it lacks T7 RNA polymerase-encoding genes and T7 promoter cannot be used for the overexpression. Then, we demonstrate that the constitutive promoter J23119 could be used for the overexpression of the red fluorescent protein (RFP)-encoding gene. Finally, the whole preparation method and transformation parameters of E. persicina competent cells are determined. Moreover, the knockout and integration system of target genes into the genome of E. persicina strains are conducted by double-crossover homologous recombination using the suicide vector. A schematic diagram of suicide plasmid–mediated genome editing is shown in Figure 1.
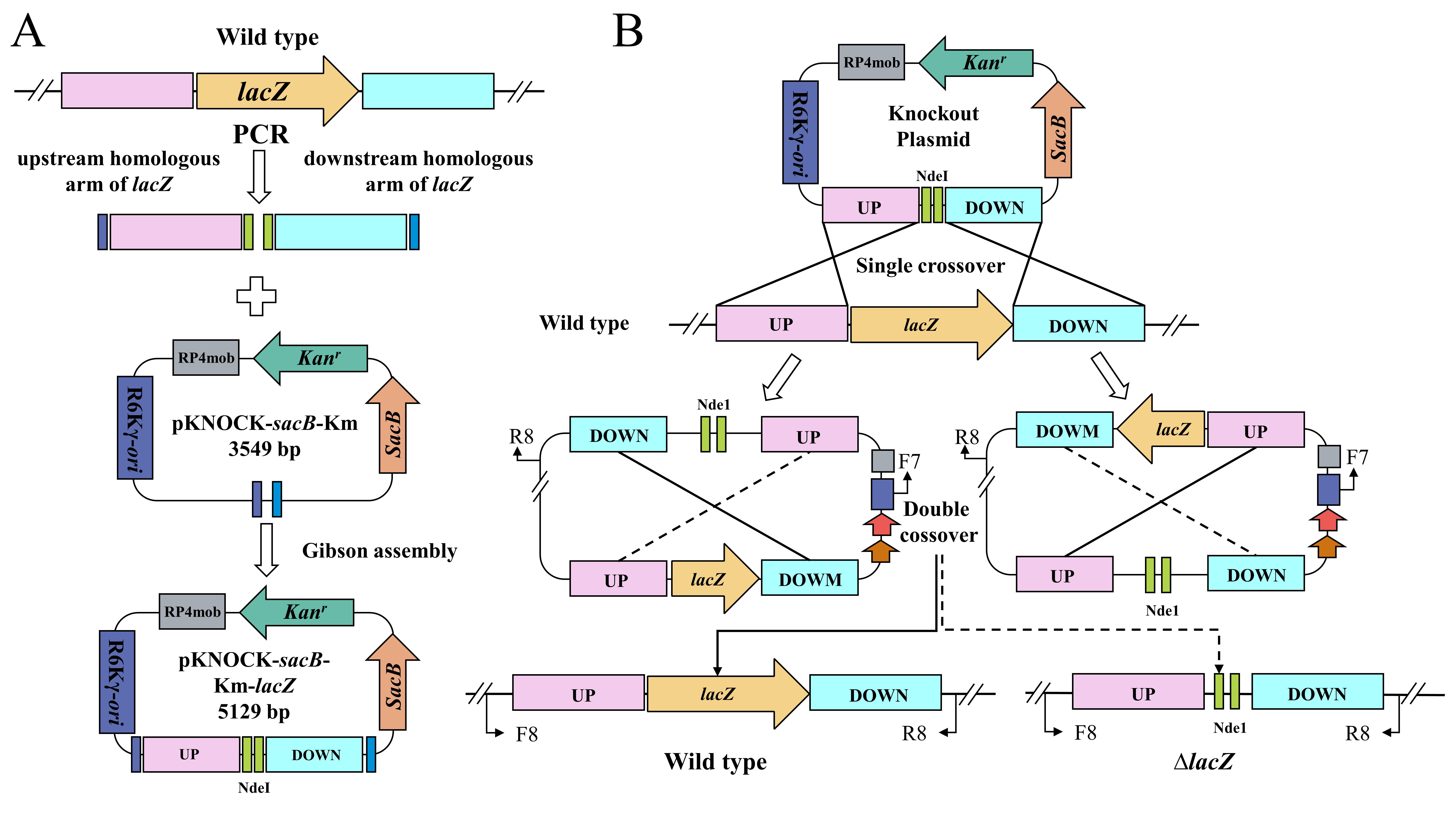
Figure 1. Schematic diagram of suicide plasmid–mediated genome editing. A. Construction of lacZ knockout plasmid. B. Parental conjugation transformation and recombination exchanges.
Materials and reagents
Biological materials
Erwinia persicina BST187 [13]
pGD plasmid [13]
pET28a-Cas9 plasmid [13]
Escherichia coli DH5α chemically competent cells (Biomed, catalog number: BC102-02)
pKNOCK-sacB-km [13]
Escherichia coli DH5α λpir chemically competent cells (ZOMANBIO, catalog number: ZK227)
Escherichia coli S17-1 λpir chemically competent cells (ANGYUBIO, catalog number: G6055-10)
Reagents
KOD OneTM PCR Master Mix (TOYOBO, catalog number: KMM-201)
Agarose (GenStar, catalog number: VA10252)
Nucleic acid dye (Biosharp, catalog number: BS354B)
StarMarker D5000 (GenStar, catalog number: M030-10)
StarMarker D8000 (GenStar, catalog number: M031-05)
New Gel Mini Purification kit (ZOMANBIO, catalog number: ZPN202-3)
EZ-HiFi Seamless Cloning kit (GenStar, catalog number: T196-20)
Super Taq DNA Polymerase (GenStar, catalog number: A012-100)
Fast Plasmid Miniprep kit (ZOMANBIO, catalog number: ZPK101-3)
Chemicals
Tryptone (OXOID, catalog number: LP0042B)
Yeast extract powder (OXOID, catalog number: LP0021B)
Agar powder (Solarbio, catalog number: A8190)
Kanamycin sulfate (Solarbio, catalog number: BS-K8020-5)
Ampicillin (Solarbio, catalog number: BS-A8180-5)
Sucrose (Sigma-Aldrich, catalog number: V900116)
NaCl (SCR, catalog number: 10019318)
KCl (SCR, catalog number: 10016318)
Na2HPO4·12H2O (Macklin, catalog number: S818120-500 g)
KH2PO4 (Macklin, catalog number: P815662-500 g)
Tris-base (Solarbio, catalog number: SJ025)
Glacial acetic acid (Macklin, catalog number: A801295-500 mL)
EDTA (Aladdin, catalog number: E116428-500 g)
CaCl2 (Macklin, catalog number: C804986-500 g)
Glycerol (DAMAO, catalog number: 2294)
Solutions
Kanamycin (50 mg/mL) (see Recipes)
Ampicillin (100 mg/mL) (see Recipes)
50% glycerol (see Recipes)
0.1 M CaCl2 (see Recipes)
0.85% NaCl (see Recipes)
50× TAE electrophoresis buffer (see Recipes)
LB medium (see Recipes)
10× Phosphate-buffered saline (PBS) (see Recipes)
Recipes
Kanamycin (50 mg/mL)
Dissolve 500 mg of kanamycin sulfate in 10 mL of ddH2O. Filter sterilize using a 0.22 µm syringe filter and store at -20 °C before use.
Ampicillin (100 mg/mL)
Dissolve 1,000 mg of ampicillin in 10 mL of ddH2O. Filter sterilize using a 0.22 µm syringe filter and store at -20 °C before use.
50% glycerol
Make the 50% glycerol solution by diluting 100% glycerol in ddH2O and autoclave.
0.1 M CaCl2
Dissolve 1.47 g of CaCl2·2H2O in ddH2O to a final volume of 100 mL and autoclave the solution.
0.85% NaCl
Dissolve 0.85 g of NaCl in ddH2O to a final volume of 100 mL and autoclave the solution.
50× TAE electrophoresis buffer (1,000 mL)
Note: Use 1 M HCl or 1 M NaOH to adjust the pH to 8.5 and dilute to 1× with ddH2O before use.
Reagent Final concentration Quantity Tris-base 2 M 242 g Glacial acetic acid 1 M 57.1 mL EDTA 100 mM 37.2 g H2O n/a To 1,000 mL LB medium (1,000 mL)
Add 1 mL of kanamycin (50 mg/mL) or ampicillin (100 mg/mL) into 1 L of LB broth or agar after autoclaving when the media has cooled to 50–55 °C.
Note: Sucrose solution should be autoclaved at 115 °C for 30 min.
Reagent Final concentration Quantity Tryptone 1% 10 g Yeast extract powder 0.5% 5 g NaCl 1% 10 g Agar powder 1.5% 15 g (agar plate) Sucrose 15% 150 g (sucrose plate) H2O n/a To 1,000 mL 10× Phosphate-buffered saline (PBS) (1,000 mL)
Use 1 M HCl or 1 M NaOH to adjust the pH to ~6.8. The pH of solution should be adjusted to 7.4 when diluted to 1× PBS.
Dilute 10× PBS to 1× PBS and then sterilize the solution by autoclaving for 15 min at 121 °C. Store it at 4 . Dissolve 1 mL of 1× PBS in 99 mL of ddH2O (0.01 M) and sterilize by autoclaving for 15 min at 121 °C before use.
Reagent Final concentration Quantity Na2HPO4·12H2O 100 mM 35.814 g KH2PO4 18 mM 2.4496 g NaCl 1.37 M 80.0669 g KCl 27 mM 2.0129 g H2O n/a To 1,000 mL
Laboratory supplies
Petri plates, 90 mm × 15 mm (Biosharp, catalog number: BS-90-D)
1.5 mL Eppendorf tubes (Biosharp, catalog number: BS-15-M)
0.2 mL PCR tubes (LABSELECT, catalog number: PST-0208-FT-C)
Pipette tips (Biosharp, catalog numbers: BS-10-T, BS-200-T, BS-1000-T)
Cell spreader (Biologix, catalog number: 65-1001)
Parafilm (Parafilm, catalog number: PM996)
12 mL Polypropylene test tube (Biosharp, catalog number: BS-PPT-12-S)
10 mL syringes (MOTUO, catalog number: MT-ZSQ-04)
Syringe filter (0.22 µm) (Merck Millipore, catalog number: C134954)
Sterile 50 mL conical tubes (LABSELECT, catalog numbers: CT-002-50A)
96-well cell culture plate (Corning, catalog numbers: CLS351172)
Blue cap reagent bottle (SHUNIU, catalog numbers: 250 mL, 500 mL)
Equipment
Pipettes (Eppendorf, model: 3121000023, 3121000090, 3120000267)
Ultrapure Water Systems (ZHIANG, model: Best-D)
PCR instruments (BIO-GENER, model: GET3XG)
Microwave oven (Midea, model: 2105210600)
DNA electrophoresis apparatus (Tanon, model: EPS-300)
Gel image system (Tanon, model: Tanon 1600R)
UV glue cutting instrument (Miulab, model: DUT-48)
Electrothermal constant temperature (MIULAB, model: DTC-100)
Water bath pot (Shanghai Yiheng, model: DK-8D)
Liquid nitrogen container (DONGYA, model: YZD-50)
Clean bench (AIRTECH, model: SW-CJ-1FD)
Ice machine (XUEKE, model: IMS-70)
Constant temperature oscillation incubator (Shanghai Zhichu, model: ZQZY-78AES)
Vortex (DLAB, model: VJ218AG0003297)
4 °C refrigerator (Zhongke Duling, model: MPC-5V656)
-20 °C ultra-low temperature freezer (Zhongke Duling, model: MDF-25V278W)
-80 °C ultra-low temperature freezer (Haier, model: DW86L626)
High-pressure sterilization pot (BioNation, model: CT-90A)
Centrifuge 5424R (Eppendorf, model: CENTRIFUGE 5424R)
High-speed freezing centrifuge (Eppendorf, model: CENTRIFUGE 5810R)
Electronic balance (RADWAG, model: WTC-2000)
Ultra-micro ultravioletvisible spectrophotometer (BIO-DL, model: Micro drop 5803032204)
Baking oven (CIMO, model: DHG-9143BS-II)
Oven incubator (CIMO, model: SPX-250BSH-II)
Microplate reader (Molecular Devices, model: SMP7)
pH meter (Sartorius, model: PB-10)
Software and datasets
SnapGene® (SnapGene, https://www.snapgene.com/)
Tanon 1600 Gel Image System (Tanon)
SoftMax Pro 7 (Molecular Devices, http://www.moleculardevices.com)
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Cheng, T., Ge, T., Zhao, X., Liu, Z. and Zhao, L. (2024). Efficient Genetic Transformation and Suicide Plasmid-mediated Genome Editing System for Non-model Microorganism Erwinia persicina. Bio-protocol 14(6): e4956. DOI: 10.21769/BioProtoc.4956.
分类
微生物学 > 微生物遗传学 > 基因组编辑
分子生物学 > DNA > 转化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link