- EN - English
- CN - 中文
Bacterial Pathogen-mediated Suppression of Host Trafficking to Lysosomes: Fluorescence Microscopy-based DQ-Red BSA Analysis
细菌病原体介导的宿主向溶酶体运输的抑制:基于荧光显微镜的DQ-Red BSA分析
发布: 2024年03月05日第14卷第5期 DOI: 10.21769/BioProtoc.4951 浏览次数: 2902

相关实验方案

HS-GC-MS 方法用于 DSS 诱导性结肠炎模型中 IBD 动态变化的诊断
Olga Yu. Shagaleeva [...] Natalya B. Zakharzhevskaya
2025年03月20日 3118 阅读
Abstract
Intracellular bacterial pathogens have evolved to be adept at manipulating host cellular function for the benefit of the pathogen, often by means of secreted virulence factors that target host pathways for modulation. The lysosomal pathway is an essential cellular response pathway to intracellular pathogens and, as such, represents a common target for bacterial-mediated evasion. Here, we describe a method to quantitatively assess bacterial pathogen–mediated suppression of host cell trafficking to lysosomes, using Salmonella enterica serovar Typhimurium infection of epithelial cells as a model. This live-cell imaging assay involves the use of a BODIPY TR-X conjugate of BSA (DQ-Red BSA) that traffics to and fluoresces in functional lysosomes. This method can be adapted to study infection with a broad array of pathogens in diverse host cell types. It is capable of being applied to identify secreted virulence factors responsible for a phenotype of interest as well as domains within the bacterial protein that are important for mediating the phenotype. Collectively, these tools can provide invaluable insight into the mechanisms of pathogenesis of a diverse array of pathogenic bacteria, with the potential to uncover virulence factors that may be suitable targets for therapeutic intervention.
Key features
• Infection-based analysis of bacterial-mediated suppression of host trafficking to lysosomes, using Salmonella enterica serovar Typhimurium infection of human epithelial cells as a model.
• Live microscopy–based analysis allows for the visualization of individually infected host cells and is amenable to phenotype quantification.
• Assay can be adapted to a broad array of pathogens and diverse host cell types.
• Assay can identify virulence factors mediating a phenotype and protein domains that mediate a phenotype.
Background
Bacterial pathogens employ diverse strategies to manipulate host cellular function to establish infection [1]. Among the many approaches, modulation of host cells can occur at the level of immune signalling and immune cell function [1,2], cellular immune responses such as lysosomal and autophagy function [3–6], organelle function [7,8], or cytoskeletal dynamics [1,9,10]. The lysosomal pathway represents a fundamental signalling hub and cellular immune response pathway in eukaryotic cells [11,12]. Lysosomes are membrane-bound organelles responsible for the degradation of proteins, lipids, nucleic acids, and carbohydrates [11,12]. These vesicles exhibit a characteristically low pH that promotes the activation of over 60 different hydrolases, ultimately contributing to cellular homeostasis and clearance of intracellular pathogens [11,12]. Defects in lysosome function can lead to one of more than 50 different disorders, collectively defined as lysosome storage diseases, with implications in neurodegeneration and aging [12–15].
Given that lysosomes function as a primary organelle responsible for the clearance of intracellular bacterial pathogens, they represent an important target for pathogen manipulation. Intracellular bacteria have evolved diverse mechanisms to manipulate host function to evade lysosomal degradation [2,10]. Lysosomal evasion can occur through either direct or indirect means. An example of an indirect mechanism of evasion is the manipulation of host trafficking to acquire membrane from other organelles. For example, the pathogen Legionella pneumophila uses a cooperative approach involving a series of secreted virulence proteins to acquire membrane from the endoplasmic reticulum [16]. This acquisition of membrane disguises the pathogen-containing vacuole as the host compartment, ultimately evading detection by the lysosomal pathway [17]. Many pathogens have evolved more direct means of targeting lysosomal function [10,18]. For example, the foodborne pathogen Vibrio parahaemolyticus uses the secreted virulence protein VepA to target the lysosomal V-ATPase and induce lysosome rupture [5].
Effective characterization of pathogen-mediated suppression of host lysosome function involves identifying the mechanism of host evasion and ultimately establishing the mechanism of action of the virulence factor responsible. This requires the development of robust cell biological assays for the study of these phenotypes. Among the most commonly proposed direct mechanisms of evasion is the suppression of trafficking of host lysosomes [10, 18]. Here, we describe a host–pathogen infection assay that can be used to investigate this phenotype [19]. This fluorescence-based microscopy assay involves the use of a BODIPY TR-X conjugate of BSA (DQ-Red BSA). This highly labelled BSA derivative exists as a self-quenched, non-fluorescent form (Figure 1A) and, upon treatment of host cells, is internalised by endocytosis. Upon trafficking to functional lysosomes, the acquisition of lysosomal proteases results in BSA cleavage, resulting in unquenching and bright-red fluorescence [20]. DQ-Red BSA is a substrate for a broad array of lysosome hydrolases and, therefore, is capable of detecting global lysosome function [21].
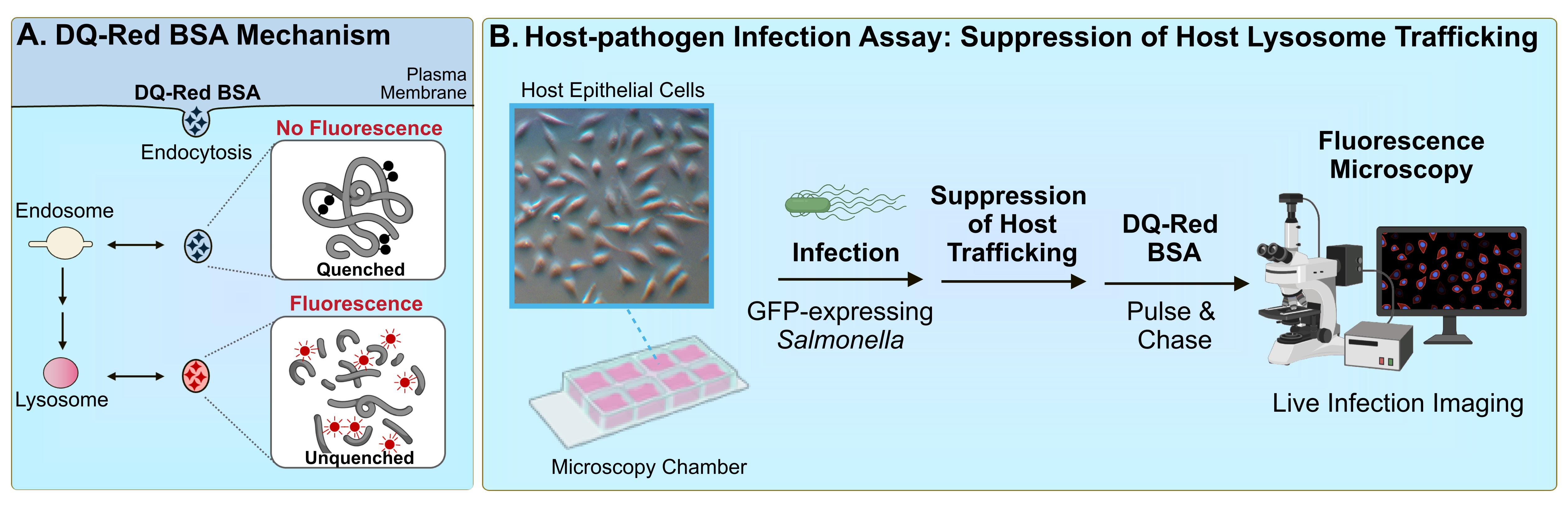
Figure 1. DQ-Red BSA assay for bacterial pathogen–mediated suppression of host trafficking. A. DQ-Red BSA dye mechanism. The fluorophores within the DQ-Red BSA dye exist in a quenched form and therefore do not generate intrinsic fluorescence. When host cells are treated with the dye, the compound is internalised by general endocytic mechanisms, and is trafficked to intermediates of the endocytic pathway. When dye-containing vesicles are trafficked to lysosomes, active lysosomal proteases cleave the BSA backbone, resulting in unquenching of the fluorophores and the generation of bright-red fluorescence. B. Host–pathogen infection assay to assess suppression of host lysosomal trafficking. Host cells are seeded in a microscopy chamber slide and infected with the bacterial pathogen of interest expressing green fluorescent protein (GFP). At the timepoint of interest, cells are subjected to a pulse-chase with DQ-Red BSA. Infected host cells are imaged by fluorescence microscopy, and suppression of host lysosome trafficking is quantified by imaging software.
The following summarises a method to quantitatively assess bacterial pathogen–mediated suppression of host cell trafficking to lysosomes, using Salmonella enterica serovar Typhimurium infection of epithelial cells as a model (Figure 1B). S. enterica is a Gram-negative pathogen that is among the leading causes of foodborne gastroenteritis worldwide [22]. As an intracellular vacuolar pathogen, S. Typhimurium evades targeting for death by lysosomes to establish a replicative niche within host cells [10]. S. Typhimurium was shown to suppress host trafficking to lysosomes (Figure 2) using this fluorescence microscopy–based imaging assay [19], which allows the visualization of trafficking to lysosomes in individual live-infected host cells alongside neighbouring non-infected host cell counterparts. This method was also used to identify the secreted virulence factor responsible for the phenotype, as well as domains within the bacterial protein that contribute to the phenotype [19]. The methods outlined highlight important considerations for adapting this method for the study of other host cell types and other bacterial pathogens, as well as for modifying the assay setup to tailor to the experimental question of interest.
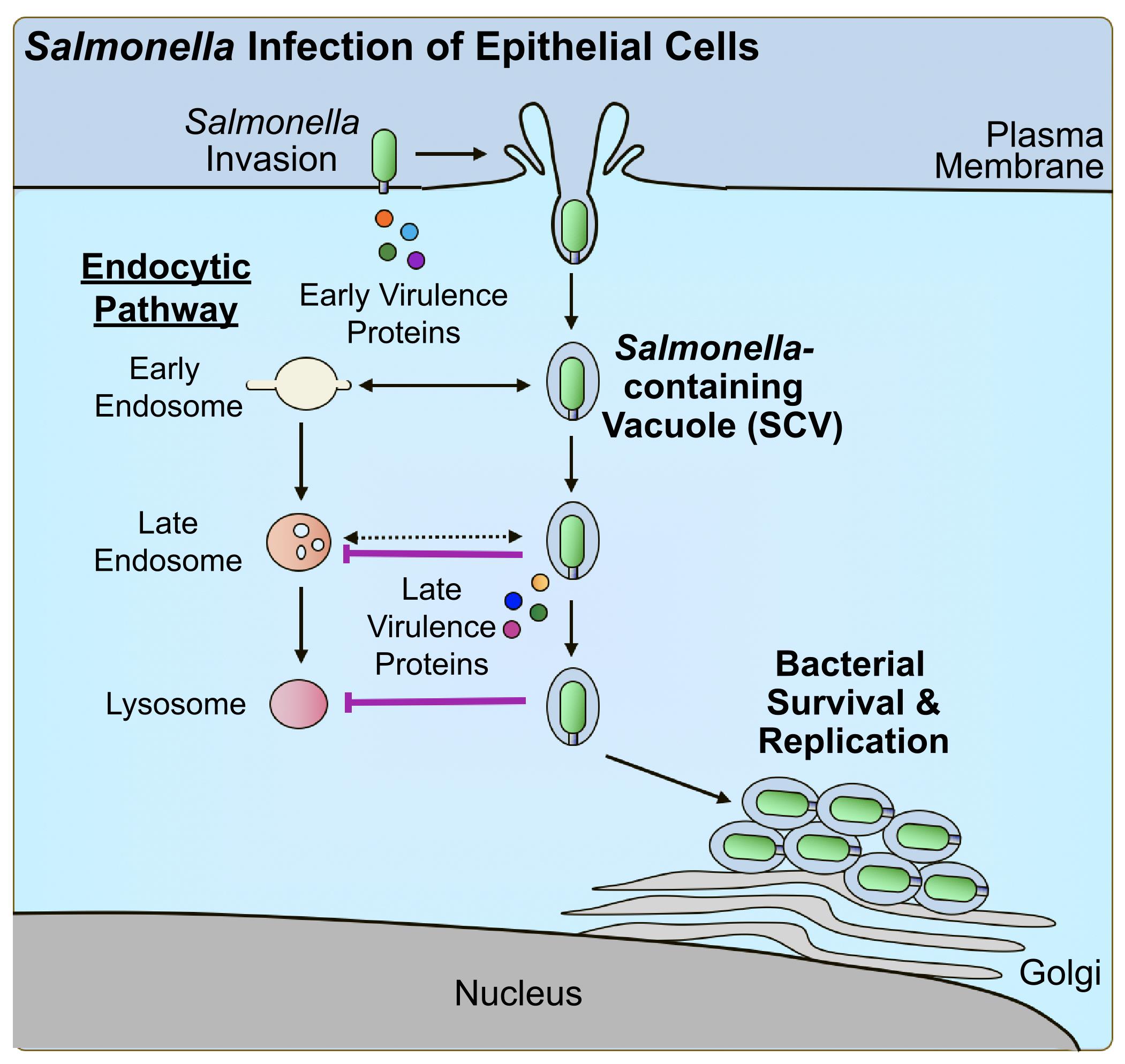
Figure 2. Salmonella virulence factor–mediated suppression of host trafficking to lysosomes. Salmonella Typhimurium manipulates the host cytoskeleton to direct its uptake into host epithelial cells using early-secreted virulence proteins. Upon uptake, Salmonella-containing vacuoles (SCVs) begin to undergo interactions with early intermediates of the endocytic pathway. As a mechanism of evasion, Salmonella produces late-secreted virulence proteins to suppress trafficking to lysosomes and promote intracellular survival and replication.
Materials and reagents
Biological materials
HeLa (ATCC, catalog number: CCL-2)
Salmonella enterica serovar Typhimurium SL1344 harbouring pFPV25.1 [23]
Note: The pathogen of interest must be expressing a fluorescent protein that is not associated with red wavelengths (e.g., GFP) for detection purposes. This may be in the form of a shuttle plasmid such as pFPV25.1 (expressing GFP) or a strain harbouring a chromosomal insertion. Should this not be previously established for the pathogen of interest, broad host spectrum shuttle plasmids exist that might be worthy of investigation [24].
Reagents
Dulbecco modified Eagle medium (DMEM), 4.5 g/L glucose, with L-glutamine, sodium pyruvate, and phenol red) (Wisent, catalog number: 319-005-CL)
Fetal bovine serum (FBS) (Wisent, catalog number: 080-450)
LB Miller broth (BioShop, catalog number: LBL407)
Agar, bacteriological grade (BioShop, catalog number: AGR001)
Streptomycin sulfate (BioShop, catalog number: STP101)
Carbenicillin sodium salt (BioShop, catalog number: CAR544)
Phosphate-buffered saline with calcium and magnesium (PBS+/+) (Wisent, catalog number: 311-011-CL)
Gentamicin sulfate (BioShop, catalog number: GTA401)
DQ-Red BSA (Thermo Fisher, catalog number: D12051)
Phosphate-buffered saline without calcium and magnesium (PBS-/-) (Wisent, catalog number: 311-425-CL)
Roswell Park Memorial Institute 1640 (RPMI), L-glutamine & HEPES, no sodium bicarbonate (Wisent, catalog number: 350-025-CL)
Solutions
Streptomycin stock solution (see Recipes)
Carbenicillin stock solution (see Recipes)
LB-agar solution (for LB-agar plates) (see Recipes)
DQ-Red BSA solution (see Recipes)
Recipes
Streptomycin stock solution (1 mL)
Note: Dissolve powder in water and vortex well until powder dissolves. In a sterile area such as a Bunsen burner or Class II biosafety cabinet, filter sterilise the stock into a sterile tube.
Reagent Final concentration Quantity or Volume Streptomycin sulfate 50 mg/mL 50 mg Double-distilled water (ddH2O) n/a to 1 mL Carbenicillin stock solution (1 mL)
Note: Dissolve powder in water and vortex well until powder dissolves. In a sterile area such as a Bunsen burner or Class II biosafety cabinet, filter sterilise stock into a sterile tube.
Reagent Final concentration Quantity or Volume Carbenicillin sodium salt 100 mg/mL 100 mg Double-distilled water (ddH2O) n/a to 1 mL LB-agar solution (500 mL)
Note: Dissolve powder in water and mix well using a stir bar until powder dissolves as much as possible. Autoclave using standard settings for liquid laboratory media. Where supplementation with antibiotics is applicable, when the autoclaved media is cooled such that it is warm to the touch, add the appropriate concentration of antibiotic stock in a sterile area. After stirring on a stir plate, use a sterile area to pour a layer of agar-supplemented media in each Petri dish. Allow to solidify at room temperature and store solidified plates at 4 °C.
Reagent Final concentration Quantity or Volume LB Miller broth (powder) 25 g/L 12.5 g Agar 15 g/L 7.5 g Double-distilled water (ddH2O) n/a to 500 mL DQ-Red BSA stock solution (1 mL)
Note: Dissolve powder in PBS-/- and mix well by vortexing. If needed, sonicate to assist with solubilizing the reagent. Once solubilised, the reagent can be stored at 4 °C for several weeks. Note that DQ-Red BSA is light-sensitive. Store in a sample tube that is either covered in foil or opaque and, wherever possible, handle in the dark.
Reagent Final concentration Quantity or Volume DQ-Red BSA 2 mg/mL 1 mg PBS-/- n/a to 500 μL
Laboratory supplies
Glass-bottom microscopy chambers (ibidi, catalog number: 80827-90)
Centrifuge tubes, polypropylene, sterile (Corning, catalog number: C352098)
Serological pipettes, sterile (Sarstedt, catalog number: 86.1253.001)
Petri dishes for bacteriology, sterile (Sarstedt, catalog number: 82.1473.001)
Round-bottom culture tubes, sterile (Corning, catalog number: C352059)
Syringe, sterile (BD, catalog number: B309659)
Syringe filter, sterile (Millipore Sigma, catalog number: SLGP033RS)
Magnetic stir bar (Fisherbrand, catalog number: 800371113)
Equipment
CO2 incubator (Nuaire, model: NU-425-400)
Bacterial incubator (New Brunswick, model: Innova 42)
Quorum spinning disk fluorescence microscope (Leica, model: DMIRE2, Hamamatsu, model: CMOS FL-400)
Bunsen burner (EISCO, model: CH0095B) or Class II biosafety cabinet (Nuaire, model: NU-602-600)
Analytical balance (Mettler Toledo, model: ML304T)
Stir plate (Thermo Scientific, model: SP131525)
Autoclave
Vortex (VWR, model: G-560)
Microcentrifuge (Eppendorf, model: 5425)
Software and datasets
Fiji (ImageJ, v1.54f 29, June 2023) (free analysis software)
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Mocăniță, M., Martz, K. and D'Costa, V. M. (2024). Bacterial Pathogen-mediated Suppression of Host Trafficking to Lysosomes: Fluorescence Microscopy-based DQ-Red BSA Analysis. Bio-protocol 14(5): e4951. DOI: 10.21769/BioProtoc.4951.
分类
微生物学 > 微生物-宿主相互作用 > 细菌
细胞生物学 > 细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link