- EN - English
- CN - 中文
Analysis of Cleavage Activity of Dengue Virus Protease by Co-transfections
通过共转染分析登革热病毒蛋白酶的切割活性
(§ Technical contact) 发布: 2024年03月05日第14卷第5期 DOI: 10.21769/BioProtoc.4946 浏览次数: 1729
评审: Luis Alberto Sánchez VargasRan ChenVaibhav B. ShahDay-Yu Chao

相关实验方案

诱导型HIV-1库削减检测(HIVRRA):用于评估外周血单个核细胞中HIV-1潜伏库清除策略毒性与效力的快速敏感方法
Jade Jansen [...] Neeltje A. Kootstra
2025年07月20日 2460 阅读
Abstract
The genome of the dengue virus codes for a single polypeptide that yields three structural and seven non-structural (NS) proteins upon post-translational modifications. Among them, NS protein-3 (NS3) possesses protease activity, involved in the processing of the self-polypeptide and in the cleavage of host proteins. Identification and analysis of such host proteins as substrates of this protease facilitate the development of specific drugs. In vitro cleavage analysis has been applied, which requires homogeneously purified components. However, the expression and purification of both S3 and erythroid differentiation regulatory factor 1 (EDRF1) are difficult and unsuccessful on many occasions. EDRF1 was identified as an interacting protein of dengue virus protease (NS3). The amino acid sequence analysis indicates the presence of NS3 cleavage sites in this protein. As EDRF1 is a high-molecular-weight (~138 kDa) protein, it is difficult to express and purify the complete protein. In this protocol, we clone the domain of the EDRF1 protein (C-terminal end) containing the cleavage site and the NS3 into two different eukaryotic expression vectors containing different tags. These recombinant vectors are co-transfected into mammalian cells. The cell lysate is subjected to SDS-PAGE followed by western blotting with anti-tag antibodies. Data suggest the disappearance of the EDRF1 band in the lane co-transfected along with NS3 protease but present in the lane transfected with only EDRF1, suggesting EDRF1 as a novel substrate of NS3 protease. This protocol is useful in identifying the substrates of viral-encoded proteases using ex vivo conditions. Further, this protocol can be used to screen anti-protease molecules.
Key features
• This protocol requires the cloning of protease and substrate into two different eukaryotic expression vectors with different tags.
• Involves the transfection and co-transfection of both the above recombinant vectors individually and together.
• Involves western blotting of the same PVDF membrane containing total proteins of the cell lysate with two different antibodies.
• Does not require purified proteins for the analysis of cleavage of any suspected substrate by the protease.
Graphical overview
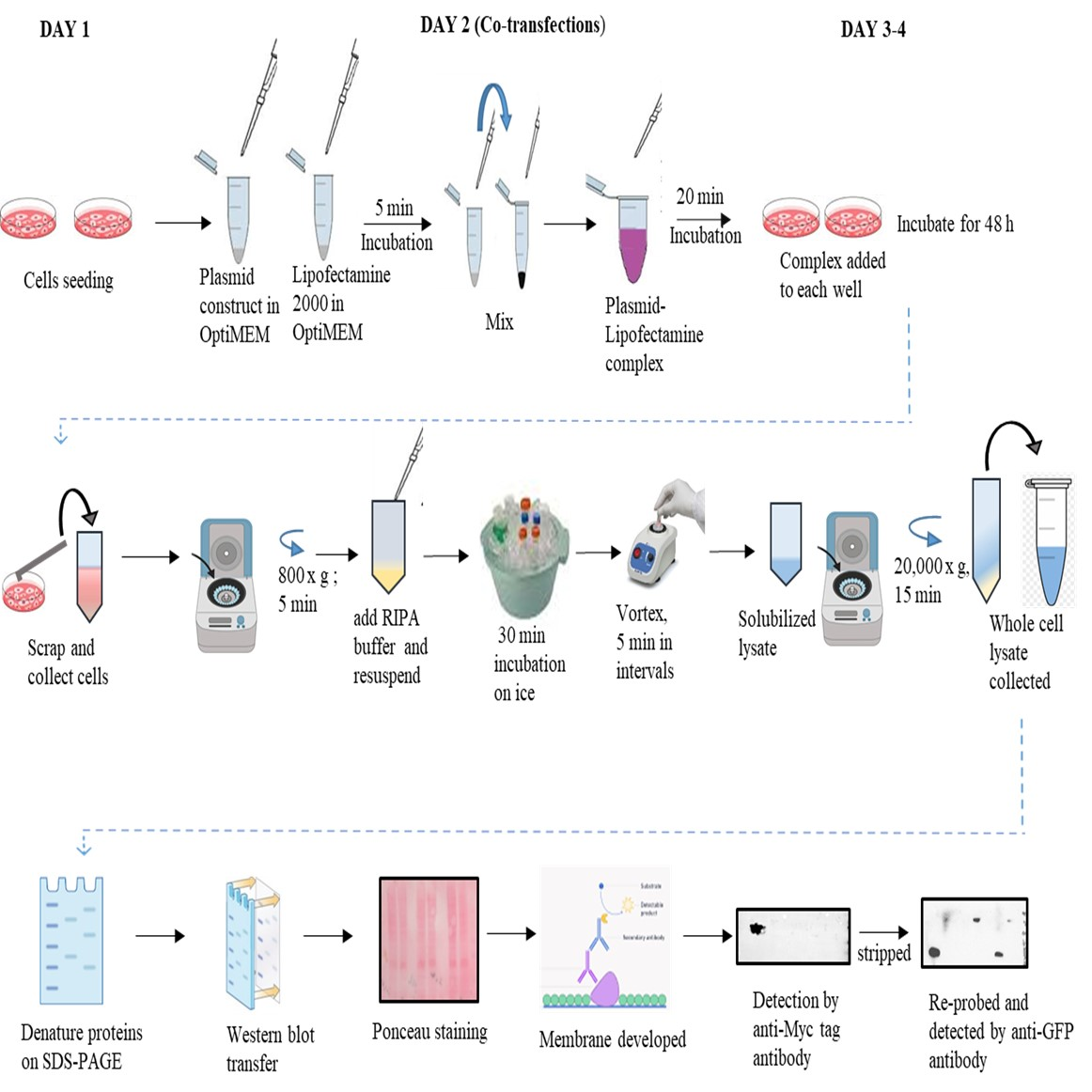
Background
Viral encoded proteins mitigate cellular activities in order to bring the host under their control. In this direction, proteases of viral origin cleave the host proteins and cause irreparable damage to the host cells, hence being prime drug targets. Poliovirus-encoded 2A protease [1] cleaves the eukaryotic initiation factor 4G (eIF4G) and stalls cellular mRNA translation. Proteases of hepatitis C virus (HCV) [2] and dengue virus [3] are known to target the mitochondrial homeostasis, which leads to mitochondrial dysfunction and failure of the immune response. The main proteases 3CLpro and the papain-like protease (PLPro) of SARS CoV2 [4] possess protease activity, and the substrates need to be characterized. For this purpose, in silico techniques are useful, but experimental evidence is needed to confirm the analysis. In vitro pull-down assays or immunoprecipitation (IP) assays are being used to identify the novel substrates of viral-encoded proteases. 2D gel electrophoresis followed by MALDI-TOFF is also being used to this purpose but often yields false positives or negatives.
Dengue virus infections are hyperendemic in more than 130 countries across the globe, causing millions of infections and thousands of deaths annually. There is no vaccine or specific drugs developed to date, in spite of several attempts. One of the reasons for this is the failure to identify suitable drug/vaccine targets. The genome of this virus encodes three structural and seven non-structural (NS) proteins, along with two untranslated regions, one at each end. Among NS proteins that play significant roles during viral replication, NS3, alone or along with NS2B, possesses a crucial role and is the prime drug target for developing antivirals. NS3 is a 70 kDa protein that contains a 180 amino acid N-terminal trypsin-like serine protease domain followed by a C-terminal helicase domain [5]. This protein is a multifunctional serine protease, forming the catalytic triad with amino acids histidine (H-51), aspartate (D-75), and serine (S-135). NS3 is also known to regulate several host proteins to induce and maintain pathogenesis. Along with cleavage of the self-polypeptide to yield functional proteins, this protease is also known to cleave the host cellular proteins FAM134B (endoplasmic receptor), Ikα/β (cellular factors), nucleoporins (Nups), MITA, MFN1, and MFN2, thereby enhancing viral replication and affecting host metabolism [6-10]. However, there is still a lack of knowledge regarding the full list of host proteins that NS3 cleaves and its biological effects, particularly with regard to pathogenicity. In vitro cleavage experiments are useful to characterize such substrates but require purified proteins. In addition, the in vitro conditions sometimes fail to mimic the natural conditions and lead to erroneous results. NS2B–NS3 interactions have been targeted for drug development [11]. Therefore, this protocol is expected to be useful in the quick screening of anti-protease molecules against NS2B–NS3 interactions. The present protocol, an ex vivo study, describes the co-transfection followed by western blotting in order to evaluate EDRF1 as a novel substrate of dengue virus protease. Furthermore, this protocol is expected to be useful in the identification of any such substrates using NS3 of any of the four serotypes of dengue virus infections.
Materials and reagents
All materials and reagents listed below can be acquired from other suppliers. In our lab, we use molecular and cell culture grade for all reagents. High-quality products are recommended for the cell culture experiments.
Biological materials
Cell lines: human embryonic kidney 293 cells (HEK 293), obtained from National Centre for Cell Science (NCCS), Pune, India
Anti-GFP (D5.1) rabbit mAB (monoclonal antibody) (Cell Signalling Technology, catalog number: 2956)
Anti-Myc tag mouse monoclonal antibody (Proteintech, catalog number: 60003-2-Ig)
Rabbit anti-mouse-IgG HRP conjugated antibody (GeNei, catalog number: 1140580011730)
Mouse anti-rabbit-IgG HRP conjugated antibody (Santa Cruz Biotechnology, catalog number: sc-2357)
Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, catalog number: 11995-065)
Fetal bovine serum (FBS) (Gibco, catalog number: 10270106)
Antibiotic (penicillin and streptomycin) (Gibco, catalog number: 15240062)
Trypsin-EDTA (1×) solution (HiMedia, catalog number: TCL007)
Lipofectamine 2000 (Invitrogen, catalog number: 11668-027)
Radio Immunoprecipitation Assay (RIPA) buffer (Sigma Aldrich, catalog number: R0278)
Femto LUCENT™ PLUS-HRP (G-Biosciences, catalog number: 786-003)
Protease inhibitor cocktail (Protease Arrest™) 100× (G-Biosciences, catalog number: 786-331)
Bradford reagent (Bio-Rad, catalog number: 5000201)
Immobilon®-P PVDF membrane (Merck Millipore, catalog number: IPVH00010)
Trypan blue (Sigma, catalog number: T6146)
OptiMEM (Gibco, catalog number: 31985062)
Ponceau S stain (Sigma, catalog number: P3504)
Tris base (HiMedia, catalog number: TC072)
Glycine (Merck, catalog number: 1.94907.0521)
Sodium dodecyl sulphate (SDS) (SR Lifesciences, catalog number: 14374)
NaCl (Merck, catalog number: 1.93206.0521)
KCl (HiMedia, catalog number: 7447-40-7)
NaH2PO4 (Merck, catalog number: MB024)
KH2PO4 (HiMedia, catalog number: PCT0009)
BSA (HiMedia, catalog number: GRM3151)
Skimmed milk powder (HiMedia, catalog number: GRM1254)
Tween-20 (SR Life Sciences, catalog number: 28599)
Absolute ethanol (Analytic CS, catalog number: 64-17-5)
Phosphate buffer saline (PBS) (Gibco, catalog number: 10010023)
GeneJet Plasmid MidiPrep kit (Thermo Scientific, catalog number: K0481)
10× PBS (see Recipes)
PBST (1× PBS and Tween-20) (see Recipes)
Blocking buffer (see Recipes)
70% ethanol (see Recipes)
10× transfer buffer (see Recipes)
Stripping buffer (see Recipes)
Recipes
10× PBS
Reagent Final concentration Quantity NaCl 1.37 M 80 g KCl 27 mM 2 g NaH2PO4
KH2PO4
100 mM
18 mM
14.4 g
2.4 g
H2O - Up to 1 L Note: Adjust pH to 7.4. Stock can be stored at 25 °C up to one month.
Note: From this 10× stock, 500 mL of 1× PBS solution can be prepared: mix 50 mL of 10× PBS with 450 mL of H2O (can be stored at 25 °C for a week). Use double-distilled water (ddH 2O) (autoclaved) for preparing 1× PBS solution.
PBST
Reagent Final concentration Quantity 10× PBS
Tween-20
H2O
1×
0.1%
n/a
50 mL
500 μL
450 mL
Note: Use ddH2O.
Blocking buffer
Reagent Final concentration Quantity BSA/skimmed milk powder
PBST
2% or 7%
1×
0.2 or 0.7 g
10 mL
Note: Prepare fresh when required. 7% blocking buffer solution is required for the blocking the PVDF membrane.
Optional: 2% blocking buffer solution is required for preparing antibody dilutions.
70% ethanol
Reagent Final concentration Quantity Ethanol
H2O
70%
n/a
350 mL
50 mL
Note: Prepare fresh when required.
10× transfer buffer
Reagent Final concentration Quantity Tris 25 mM 30.2 g Glycine 192 mM 144 g H2O - up to 1 L Note: From this 10× stock, 1× transfer buffer can be prepared: dissolve 100 mL of 10× transfer buffer in 200 mL of methanol and make up the volume with water up to 1 L. Store at 4 °C; this can be reused for two weeks.
Stripping buffer
Reagent Final concentration Quantity Glycine 199 mM 1.5 g SDS 0.1% 0.1 g Tween-20 1% 1 mL H2O up to 100 mL Note: Adjust to pH 2.2.
Laboratory supplies
Polypropylene conical-bottom centrifuge tubes, 15 and 50 mL Falcon (Tarsons, catalog number: 546021 and 546041) (sterile)
T-25 cell culture flasks (Tarsons, catalog number: 950040)
60 mm cell culture dishes (Tarsons, catalog number: 960020)
Pipettes (1000, 200, and 20 μL) (Eppendorf, catalog numbers: 3123000063, 3123000055, 3123000098) and tips (Tarsons, catalog numbers: 523104, 523101, 523109)
Hand gloves (Kimberlay Gloves, catalog number: KC500-S)
1.5 mL Eppendorf tubes (Tarsons, catalog number: 500010) and CRYOCHILL™ Vial 2D Coded (Tarsons, catalog number: 883192)
Parafilm 4" × 125' (SR Lifesciences, catalog number: 000814)
Aluminum foil
Discard box (any supplier)
Spray bottles (any supplier)
Cell scrapper (Tarsons, catalog number: 960051)
Equipment
-80 °C deep freezer (Eppendorf, catalog number: F570)
Microcentrifuge (Eppendorf, catalog number: 5424R)
CO2 incubator (Eppendorf, catalog number: Galaxy 48R)
Inverted brightfield microscope (Lawrence and Mayo, catalog number: TC5400)
Chemidoc image analyzer (Bio-Rad, catalog number: 12003028)
Electroblot transfer unit (BioNova)
SDS-PAGE unit (Bio-Rad, model: Mini-PROTEAN Tetra Cell, catalog number: 1658038)
Hemocytometer (Neubeur Blood Counting Chamber)
Laminar hood (Laminar Flow Systems)
Animal cell culture facility
Autoclave (KETAN, catalog number: PA21)
Water bath (GeNei, catalog number: 107931GB)
Vortex mixer (Tarsons, catalog number: 3002)
Software and datasets
Image Lab software (Bio-Rad) (v6.1.0, 2020)
Zen software for Carl Zeiss fluorescence microscope (Zen 2.3, blue edition, v2.3.69.1000)
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Gandhi, L. and Venkataramana, M. (2024). Analysis of Cleavage Activity of Dengue Virus Protease by Co-transfections. Bio-protocol 14(5): e4946. DOI: 10.21769/BioProtoc.4946.
分类
微生物学 > 微生物-宿主相互作用 > 病毒
细胞生物学 > 基于细胞的分析方法 > 酶学测定
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










