- EN - English
- CN - 中文
Quantitative Determination of Cholesterol Hydroxylase Specificities by GC–MS/MS in Living Mammalian Cells
通过 GC-MS/MS 在哺乳动物活细胞中定量测定胆固醇羟化酶的特异性
发布: 2024年01月20日第14卷第2期 DOI: 10.21769/BioProtoc.4924 浏览次数: 2350
评审: Faraz RashidSuprabhat MukherjeeAnonymous reviewer(s)

相关实验方案
使用多重反应监测(MRM)分析非酒精性脂肪性肝炎(NASH)小鼠肝匀浆中脂质分布的脂质组学工作流程
Hai Ning Wee [...] Jianhong Ching
2023年07月05日 2426 阅读
Abstract
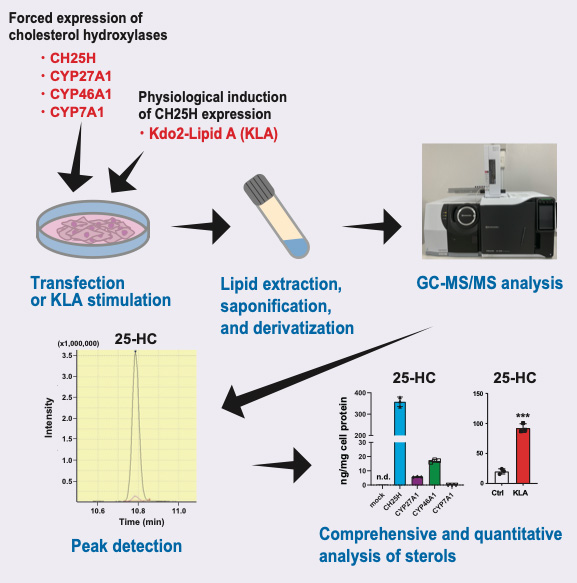
Cholesterol is oxygenated by a variety of cholesterol hydroxylases; oxysterols play diverse important roles in physiological and pathophysiological conditions by regulating several transcription factors and cell-surface receptors. Each oxysterol has distinct and overlapping functions. The expression of cholesterol hydroxylases is highly regulated, but their physiological and pathophysiological roles are not fully understood. Although the activity of cholesterol hydroxylases has been characterized biochemically using radiolabeled cholesterol as the substrate, their specificities remain to be comprehensively determined quantitatively. To better understand their roles, a highly sensitive method to measure the amount of various oxysterols synthesized by cholesterol hydroxylases in living mammalian cells is required. Our method described here, with gas chromatography coupled with tandem mass spectrometry (GC–MS/MS), can quantitatively determine a series of oxysterols endogenously synthesized by forced expression of one of the four major cholesterol hydroxylases—CH25H, CYP7A1, CYP27A1, and CYP46A1—or induction of CH25H expression by a physiological stimulus. This protocol can also simultaneously measure the amount of intermediate sterols, which serve as markers for cellular cholesterol synthesis activity.
Key features
• Allows measuring the amount of a variety of oxysterols synthesized endogenously by cholesterol hydroxylases using GC–MS/MS.
• Comprehensive and quantitative analysis of cholesterol hydroxylase specificities in living mammalian cells.
• Simultaneous quantification of intermediate sterols to assess cholesterol synthesis activity.
Graphical overview
Background
Cholesterol plays diverse important biological roles, including function regulation of biological membranes and membrane proteins, and is the precursor for steroid hormones and bile acids; thus, cellular cholesterol homeostasis is tightly controlled by multiple mechanisms (Chang et al., 2006; Yamauchi and Rogers, 2018; Luo et al., 2020). Mammalian cells cannot break down the sterol backbone. Instead, cholesterol is converted to cholesteryl ester for storage (Chang et al., 2006) and to various oxysterols (Mutemberezi et al., 2016). Oxysterols exert different functions depending on the hydroxylation site(s). Multiple oxysterols whose side chain is hydroxylated, including 25-hydroxycholesterol (25-HC) and 27-HC, regulate cellular cholesterol homeostasis; they modulate two important transcription factors: sterol regulatory element-binding protein-2 (SREBP-2) and liver X receptor (LXR) (Gill et al., 2008; M.S. Brown et al., 2018). SREBP-2 transactivates most genes involved in cholesterol acquisition (biosynthesis and uptake) (Horton et al., 2002). On the other hand, LXR upregulates the expression of several ATP-binding cassette (ABC) transporters including ABCA1 and ABCG1 that mediate the export of excess cellular cholesterol (Tontonoz, 2011). Mechanistically, side-chain oxysterols bind to Insig-1 and Insig-2—retention factors for SREBP-2 in the endoplasmic reticulum (ER)—and protect them from proteasomal degradation, thereby inhibiting SREBP-2 activation. Side-chain oxysterols also serve as natural LXR ligands when added exogenously.
A series of hydroxylases catalyze the site-specific hydroxylation of cholesterol (Mutemberezi et al., 2016). Although a variety of oxysterols have been identified in the body, 7α-HC, 27-HC, 24S-HC, and 25-HC are the most abundant. These four oxysterols are synthesized largely by CYP7A1, CYP27A1, CYP46A1, and CH25H, respectively, in the ER or mitochondria (Figure 1), while some of these hydroxylases also produce other oxysterols as minor products (Mutemberezi et al., 2016; Saito et al., 2023). Recent studies show that the expression of cholesterol hydroxylases is highly regulated in physiological and pathophysiological conditions (Cyster et al., 2014; A.J. Brown et al., 2021). CH25H expression and 25-HC biosynthesis are markedly upregulated upon infection in immune cells such as macrophages, and 25-HC itself exhibits anti-bacterial and anti-viral effects, protecting cells from infection (Bauman et al., 2009; S.Y. Liu et al., 2013; Zhou et al., 2020). 25-HC can be further hydroxylated at the 7-position by the hydroxylase CYP7B1, generating 7α,25-dihydroxycholesterol (7α,25-diHC). This dihydroxycholesterol is a ligand for the G-protein coupled receptor GPR183 (also known as EBI2) involved in immune cell migration (C. Liu et al., 2011). Higher circulating 27-HC levels associate with the risk of estrogen receptor-positive breast cancer, since 27-HC serves as an endogenous selective estrogen receptor modulator (Nelson et al., 2013). CYP46A1 is a neuron-specific cholesterol hydroxylase that converts cholesterol into 24S-HC for eliminating excess cholesterol in the brain (Russell et al., 2009). CYP7A1 is a liver-specific hydroxylase that serves as the rate-limiting enzyme for bile acid synthesis (Russell, 2009). Although the activity of these cholesterol hydroxylases is biochemically studied using radiolabeled cholesterol as the substrate, comprehensive and quantitative characterization of their specificities remains poorly explored in living cells. Furthermore, precise measurement of the products of these cholesterol hydroxylases is crucial for better understanding their physiological and pathophysiological roles.
Here, we provide a highly sensitive method using gas chromatography coupled with tandem mass spectrometry (GC–MS/MS) to determine various oxysterols synthesized in living mammalian cells where a cholesterol hydroxylase is forcedly expressed or is upregulated by lipopolysaccharide, a component of Gram-negative bacteria known to upregulate CH25H expression in macrophages (Bauman et al., 2009). The current GC–MS/MS protocol enabled us to reveal cholesterol hydroxylase–specific production of oxysterols (Saito et al., 2023). Our protocol can also simultaneously determine the amount of intermediate sterols to monitor the activity of cholesterol biosynthesis. Accordingly, our GC–MS/MS-based sterol analysis, in combination with biochemical and gene expression studies, has suggested that side-chain oxysterols enzymatically synthesized within cells primarily regulate SREBP-2 but not LXR (Saito et al., 2023).
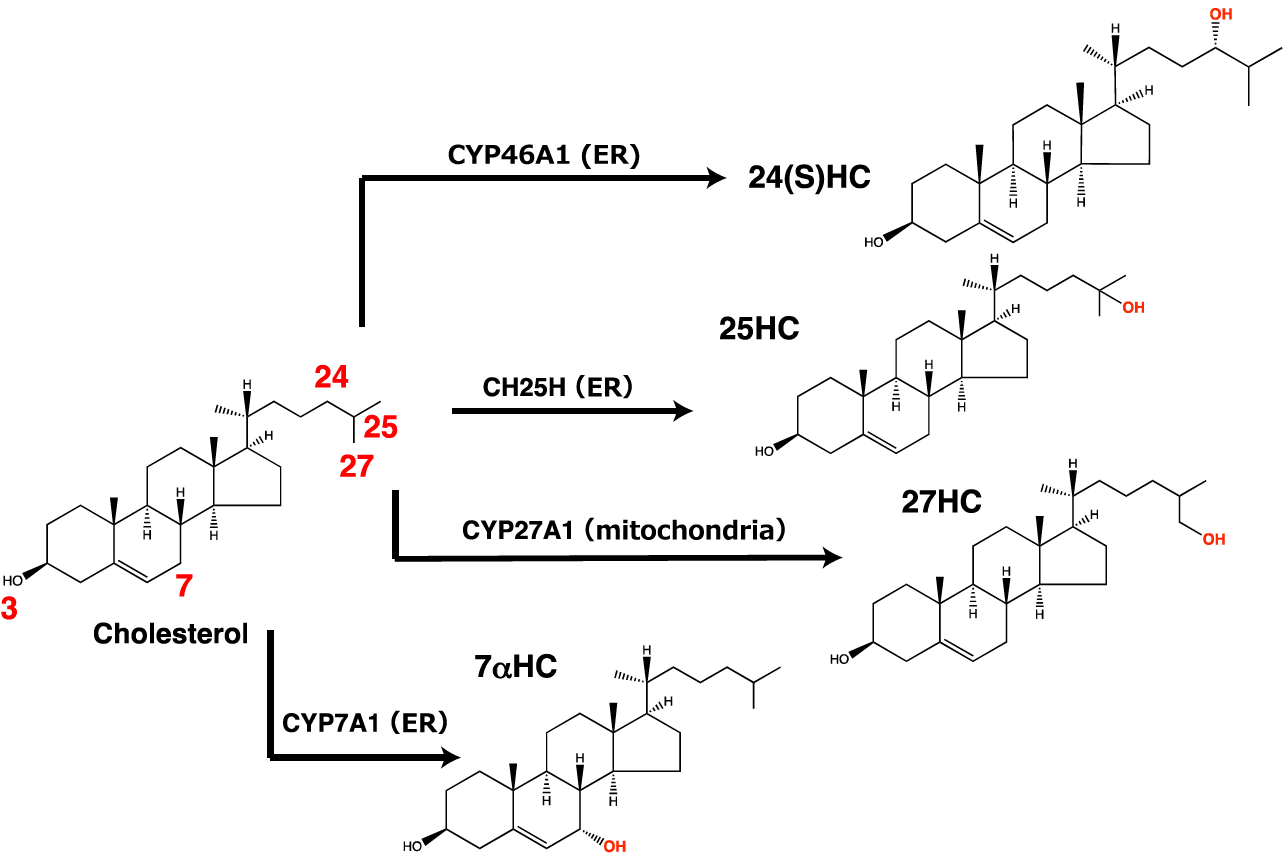
Figure 1. Hydroxylation of cholesterol by the four cholesterol hydroxylases handled in this protocol. CH25H, CYP27A1, CYP46A1, and CYP7A1 mainly produce 25-HC, 27-HC, 24(S)-HC, and 7α-HC, respectively.
Materials and reagents
1.5 mL sampling tubes (WATSON, catalog number: 131-8155C)
Screw-top test tube (Maruemu, catalog number: NR-10)
Mighty Vials (Maruemu, catalog number: 84-0561)
TORAST vial (Shimadzu GLC, catalog number: GLCTV-902)
TORAST vial insert (Shimadzu GLC, catalog number: GLCTV-I04)
TORAST vial cap (Shimadzu GLC, catalog number: GLCTV-903)
Hexane (Fuji Film Wako, catalog number: 085-00416)
2-propanol (Fuji Film Wako, catalog number: 166-04836)
Dibutyl hydroxytoluene (BHT) (Fuji Film Wako, catalog number: 029-07392)
Nitrogen gas
Helium gas
Argon gas
Chloroform (Fuji Film Wako, catalog number: 038-02606)
Ethanol (Fuji Film Wako, catalog number: 057-00456)
Milli-Q water
Pyridine (Fuji Film Wako, catalog number: 161-18453)
Note: Store pyridine in a desiccator with silica gel at room temperature and use within four weeks after opening.
N-methyl-N-trimethylsilyl trifluoroacetamide (MSTFA) (GL Science, catalog number: 1022-11061)
Cholesterol (purity ≥ 99%) (Sigma-Aldrich, catalog number: C8667)
Cholesterol-d7 (purity ≥ 99%) (Avanti Polar Lipids, catalog number: 700041P)
Desmosterol (purity ≥ 99%) (Nagara Science, catalog number: NS460402)
Lanosterol (purity ≥ 99.5%) (Nagara Science, catalog number: NS460102)
Lathosterol (purity ≥ 99%) (Nagara Science, catalog number: NS460502)
7-dehydrocholesterol (purity ≥ 95%) (Sigma-Aldrich, catalog number: 30800)
24,25-epoxycholesterol (purity ≥ 95%) (Abcam, catalog number: Ab141633)
24,25-dihydrolanosterol (purity ≥ 99.5%) (Nagara Science, catalog number: NS460201)
4β-hydroxycholesterol (purity ≥ 95%) (Cayman, catalog number: 19518)
7α-hydroxycholesterol (purity ≥ 99%) (Avanti Polar Lipids, catalog number: 700034P)
7β-hydroxycholesterol (purity ≥ 95%) (Sigma-Aldrich, catalog number: H6891)
7α,25-dihydroxycholesterol (purity ≥ 98%) (Sigma-Aldrich, catalog number: SML0541)
7α,27-dihydroxycholesterol (purity ≥ 99%) (Avanti Polar Lipids, catalog number: 700024P)
25-hydroxycholesterol (purity ≥ 98%) (Sigma-Aldrich, catalog number: H1015)
24(S)-hydroxycholesterol (purity ≥ 98%) (Sigma-Aldrich, catalog number: SML1648)
25-hydroxycholesterol-d6 (Avanti Polar Lipids, catalog number: LM4113-1EA)
27-hydroxycholesterol (purity ≥ 98%) (Sigma-Aldrich, catalog number: SML2042)
Sodium chloride (Fuji Film Wako, catalog number: 196-01665)
Potassium chloride (SIGMA, catalog number: P9541)
Disodium hydrogen phosphate dodecahydrate (Fuji Film Wako, catalog number: 196-02835)
Potassium dihydrogen phosphate (Fuji Film Wako, catalog number: 196-04245)
Potassium hydroxide (Fuji Film Wako, catalog number: 168-21815)
Sodium hydroxide (Fuji Film Wako, catalog number: 192-15985)
D-MEM/Ham’s F-12 with L-Glutamine and Phenol Red (Fuji Film Wako, catalog number: 048-29785)
RPMI-1640 with L-Glutamine and Phenol Red (Fuji Film Wako, catalog number: 189-02025)
Opti-MEMTM I reduced serum medium (Thermo Fisher Scientific, catalog number: 31985070)
Fetal bovine serum (FBS) (lot number: 27419002) (Corning, catalog number: 35-079-CF)
Penicillin G potassium (Meiji Seika Pharma, catalog number: 6111400D3051)
Streptomycin sulfate (Meiji Seika Pharma, catalog number: 6161400D1034)
Lipofectamine LTX reagent (Thermo Fisher Scientific, catalog number: 15338100)
Doxycycline hyclate (LKT Labs, catalog number: D5897)
Kdo2-Lipid A (Avanti Polar Lipids, catalog number: 69500P)
Pierce BCA Protein Assay kit (Thermo Fisher Scientific, catalog number: 23227)
Biological materials
CHO-K1 cells (gift from Dr. Ta-Yuan Chang, Geisel School of Medicine at Dartmouth)
CHO-CH25Htet-on cells (Saito et al., 2023)
J774.1 cells (RIKEN Cell Bank, catalog number: RCB0434)
Expression plasmids encoding cholesterol hydroxylase (Saito et al., 2023): pFLAG-CH25H, pCYP27A1-FLAG, pCYP46A1-FLAG, and pCYP7A1-FLAG
Note: Detailed information on CHO-CH25Htet-on cells and the four hydroxylase expression plasmids are described in the original paper (Saito et al., 2023).
Solutions
70% Ethanol (see Recipes)
75% Ethanol (see Recipes)
10 N KOH (dissolved in 75% ethanol) (see Recipes)
Hexane/2-propanol (3:2) with 0.01% BHT (see Recipes)
0.1 N NaOH (see Recipes)
PBS (see Recipes)
Recipes
70% Ethanol
Reagent Volume Ethanol (absolute) 35 mL H2O 15 mL Total 50 mL 75% Ethanol
Reagent Volume Ethanol (absolute) 37.5 mL H2O 12.5 mL Total 50 mL 10 N KOH (dissolved in 75% ethanol)
Reagent Quantity Potassium hydroxide 28.1 g 75% Ethanol up to 50 mL Total 50 mL Hexane/2-propanol (3:2) with 0.01% BHT
Reagent Quantity Hexane 60 mL 2-propanol 40 mL Dibutyl hydroxytoluene 10 mg Total 100 mL 0.1 N NaOH
Reagent Quantity Sodium hydroxide 0.2 g H2O up to 50 mL Total 50 mL PBS
Reagent Quantity Sodium chloride 8 g Potassium chloride 0.2 g Disodium hydrogen phosphate dodecahydrate 3.6 g Potassium dihydrogen phosphate g H2O up to 1 L Sodium hydroxide 1 L
Equipment
GC–MS/MS (Shimadzu, model: GCMS-TQ8040NX) equipped with an auto-sampler (AOC-20i)
BPX5 GC capillary column (30 m × 0.25 mm, 0.25 μm) (TRAJAN, catalog number: SGE-054101)
Rxi Guard column (5 m × 0.25 mm) (Restek, catalog number: 10029)
Eppendorf ThermoMixer C (Eppendorf, catalog number: 5382000023)
Hamilton micro syringe 710RN (Hamilton, catalog number: 72-5004)
Pressured gas blowing concentrator (EYELA, model: MGS-2200)
Aluminum block MGB-1540 (EYELA, catalog number: 207580)
Aluminum block MGB-1624 (EYELA, catalog number: 207610)
Low-speed refrigerated centrifuge (TOMY, model: AX-511)
SpectraMax (Molecular Devices, model: M2e)
Software and datasets
GC-MSsolution v4 (Shimadzu)
RStudio software (Posit)
GraphPad Prism 9 (GraphPad)
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Saito, H., Nishimura, M., Sato, R. and Yamauchi, Y. (2024). Quantitative Determination of Cholesterol Hydroxylase Specificities by GC–MS/MS in Living Mammalian Cells. Bio-protocol 14(2): e4924. DOI: 10.21769/BioProtoc.4924.
- Saito, H., Tachiura, W., Nishimura, M., Shimizu, M., Sato, R. and Yamauchi, Y. (2023). Hydroxylation site-specific and production-dependent effects of endogenous oxysterols on cholesterol homeostasis: Implications for SREBP-2 and LXR. J Biol Chem 299(1): 102733.
分类
生物化学 > 脂质 > 脂质测定
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link