- EN - English
- CN - 中文
Methodology to Create Auxin-Inducible Degron Tagging System to Control Expression of a Target Protein in Mammalian Cell Lines
创建生长素诱导降解子标记系统以控制哺乳动物细胞系中靶蛋白表达的方法
发布: 2024年01月20日第14卷第2期 DOI: 10.21769/BioProtoc.4923 浏览次数: 3759
评审: Rajesh RanjanBhuvanasundar RanganathanShashi Kumar Suman
Abstract
The auxin-inducible degron (AID) system is a versatile tool in cell biology and genetics, enabling conditional protein regulation through auxin-induced degradation. Integrating CRISPR/Cas9 with AID expedites tagging and depletion of a required protein in human and mouse cells. The mechanism of AID involves interactions between receptors like TIR1 and the AID tag fused to the target protein. The presence of auxin triggers protein ubiquitination, leading to proteasome-mediated degradation. We have used AID to explore the mitotic functions of the replication licensing protein CDT1. Swift CDT1 degradation via AID upon auxin addition achieves precise mitotic inhibition, revealing defects in mitotic spindle structure and chromosome misalignment. Using live imaging, we found that mitosis-specific degradation of CDT1 delayed progression and chromosome mis-segregation. AID-mediated CDT1 inhibition surpasses siRNA-based methods, offering a robust approach to probe CDT1’s mitotic roles. The advantages of AID include targeted degradation and temporal control, facilitating rapid induction and reversal of degradation—contrasting siRNA’s delayed RNA degradation and protein turnover. In summary, the AID technique enhances precision, control, and efficiency in studying protein function and regulation across diverse cellular contexts. In this article, we provide a step-by-step methodology for generating an efficient AID-tagging system, keeping in mind the important considerations that need to be adopted to use it for investigating or characterizing protein function in a temporally controlled manner.
Key features
• The auxin-inducible degron (AID) system serves as a versatile tool, enabling conditional protein regulation through auxin-induced degradation in cell biology and genetics.
• Integration of CRISPR/Cas9 knock-in technology with AID expedites the tagging and depletion of essential proteins in mammalian cells.
• AID’s application extends to exploring the mitotic functions of the replication licensing protein CDT1, achieving precise mitotic inhibition and revealing spindle defects and chromosome misalignment.
• The AID system and its diverse applications advance the understanding of protein function and cellular processes, contributing to the study of protein regulation and function.
Graphical overview
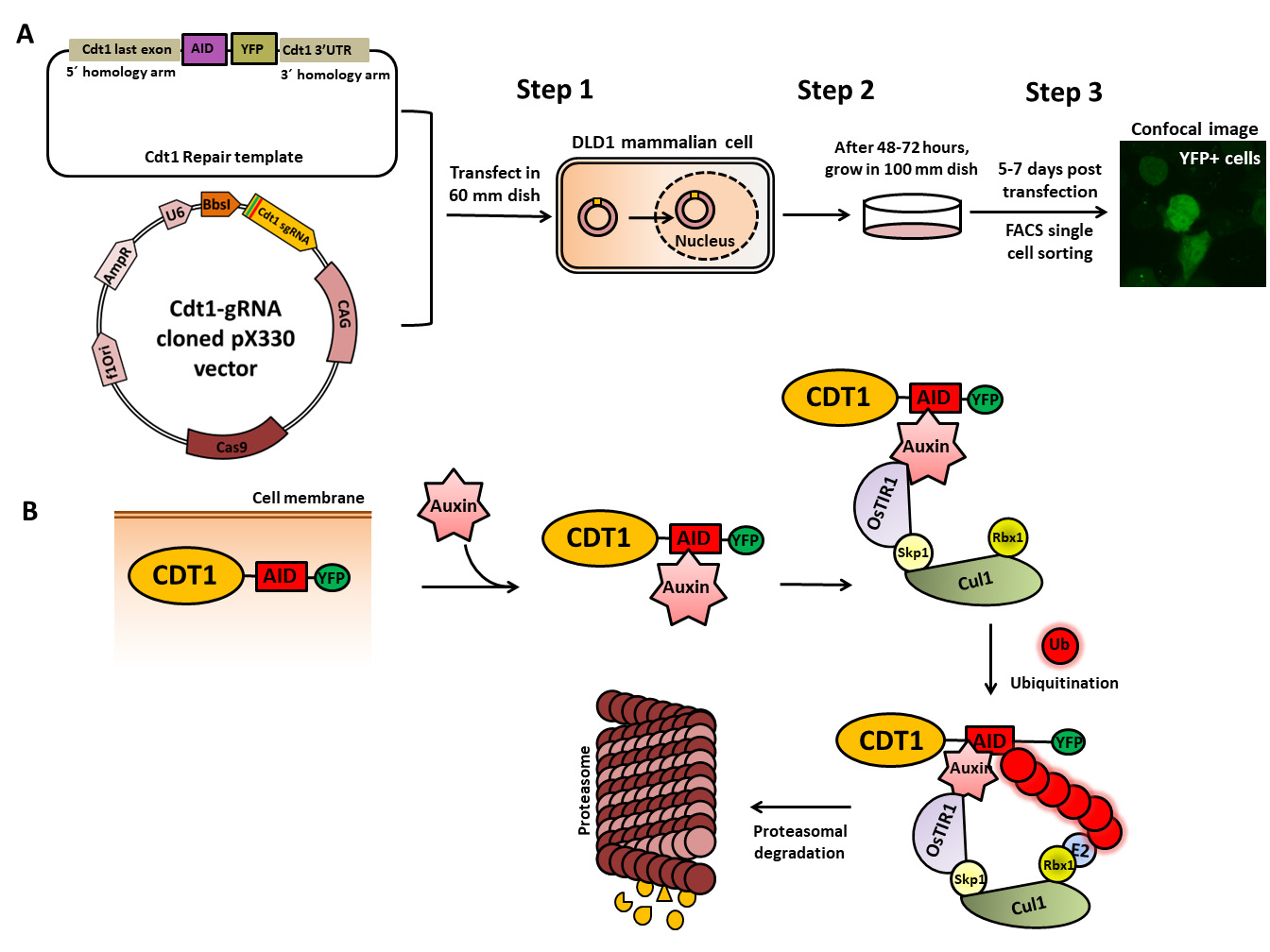
Cdt1–auxin-inducible degron (AID) tagging workflow. (A) Schematic of the cloned Cdt1 gRNA vector and the repair template generated to endogenously tag the Cdt1 genomic locus with YFP and AID at the C-terminal using CRISPR/CAS9-based genome editing. The two plasmids are transfected into DLD1-TIR1 stable cells, followed by sorting and scaling up of YFP-positive single cells. (B) The molecular mechanism of auxin-induced proteasome-mediated degradation of the target protein (CDT1) shown at the bottom of the figure is well worked out.
Background
Conditional protein degradation is an invaluable approach to understand cellular function. Among the various techniques available, the auxin-inducible degron (AID) system stands out. AID is a versatile molecular tool extensively utilized in cell biology and molecular genetics for the conditional destabilization of target proteins, facilitated by CRISPR/Cas9 gene knock-in technology [1, 2]. This mechanism capitalizes on the unique ability of the plant hormone auxin to rapidly degrade specific proteins bearing an AID sequence not only in non-plant cells like DLD1, HCT116, and HeLa but also in organisms like Caenorhabditis elegans, mouse, and yeast [3–6]. The AID assay furnishes a valuable means for probing protein functions and regulating protein expression in live cells. Its precise control over protein degradation addresses limitations of other methods such as RNAi gene silencing, which is both slow and less specific, lacking conditionality. The AID system, on the other hand, operates at the protein level, offering simplicity, rapidity, effectiveness, and reversibility. Central to this system is the interaction between the transport inhibitor response 1 receptor (TIR1), an F-box related protein, and the AID tag (available as 25 or 7 kDa), genetically fused to the protein of interest [7–9]. The half-life degradation of the tag, which is approximately 30 min, can be observed in a mammalian cell line expressing TIR1. Upon auxin binding to the TIR1 receptor, a conformational change occurs, allowing TIR1 to bind to the AID tag on the target protein. This interaction triggers the activation of the SCF (Skp1-Cullin-F-box) E3 ubiquitin ligase complex, comprising Skp1, Cul1, and RBX1. RBX1 associates with the E2 ubiquitin ligase, facilitating the ubiquitination of the protein of interest and its subsequent proteasomal degradation (26S proteasome). This system is advantageous as it leads to rapid degradation of the protein of interest in the presence of auxin, and its removal restores the phenotype [10, 11].
In our study, we employed the AID system as an innovative approach to investigate the mitotic functions of the replication licensing protein, CDT1. Through the AID system, we achieved precise and controlled inhibition of CDT1 during mitosis by inducing rapid degradation through auxin addition [12]. This strategy unveiled significant defects in mitotic spindle structure and chromosome alignment in treated cells. The system’s simplicity, control, and reversibility render it highly valuable for studying essential proteins and unraveling their roles in various cellular processes.
Materials and reagents
Cas9-gRNA expression vector (e.g., pX330, PX458, PX459)
Parent repair template (a gift from Dan Foltz Lab)
Sense oligo for cloning of sg at BbsI site (5'-CACCG [sgRNA Target Sequence]-3')
Antisense oligo for cloning at BbsI site (5'-AAAC [reverse complement sgRNA Target Sequence] C-3')
Biological materials
DLD1-TIR1 cells, colorectal adenocarcinoma cell line (a kind gift from Andrew Holland, Johns Hopkins University, Baltimore, MD, USA)
One ShotTM Stbl3TM chemically competent E. coli (Thermo Fisher Scientific, catalog number: C737303)
Reagents
Dulbecco's modified Eagle's medium (DMEM) (Corning, catalog number: 10-013-CV)
Penicillin-Streptomycin 10,000 U/mL (Thermo Fisher Scientific, catalog number: 15140122)
Trypsin-EDTA, 1× (CORNING, catalog number: 25-052-CI)
Phosphate-buffered saline (PBS 1×) (CORNING, catalog number: 21-040-CV)
Effectene transfection agent (Qiagen, catalog number: 301425)
Dimethyl sulfoxide (DMSO) (Fisher Bio ReagentsTM, catalog number: BP231-100)
Auxin (Indole-3-acetic acid, IAA) (Sigma-Aldrich, catalog number: 15148-2G)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906)
Anti-GFP mouse monoclonal (Thermo Fisher Scientific, catalog number: A-11120)
Anti-Cdt1 rabbit polyclonal H-300 (Santa Cruz Biotech, catalog number: sc-28262)
Anti-Tubulin anti-mouse monoclonal DM1A (Santa Cruz Biotech, catalog number: sc-32293)
HRP-conjugated rabbit/mouse antibodies (Azure Biosystems, catalog numbers: AC2114 and AC2115)
SuperSignalTM West Pico PLUS Chemiluminescent Substrate (Thermo Scientific, catalog number: 34580)
Plasmid Isolation kit (Qiagen, catalog number: 27104)
Gel Extraction kit (Plasmid and PCR clean-up kit) (Qiagen, catalog number: 28704)
Quick-DNA Microprep kit (Zymo Research, catalog number: D3020)
PCR Master Mix (2×) reagents (Thermo Scientific, catalog number: K0171)
GeneRuler 1 kb DNA ladder (Thermo Scientific, catalog number: SM0311)
TriTrack DNA loading dye (6×) (Thermo Scientific, catalog number: R1161)
T4 DNA ligase enzyme (NEB, catalog number: M0202S)
10× buffer for T4 DNA ligase (NEB, catalog number: B0202S)
Bbs1 enzyme (NEB, catalog number: R0539S)
Ethidium bromide (Bio-Rad, catalog number: 161-0433)
Agarose HS (Denville Scientific Inc., catalog number: CA3510-8)
Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: 03620)
Sodium dodecyl sulphate (SDS) (Fisher Scientific, catalog number: BP166-500)
Tris-base (ChemCruz, catalog number: 77-86-1)
Ammonium persulfate (APS) (Sigma-Aldrich, catalog number: A3678)
TEMED (GE Healthcare, catalog number: 17-1312-01)
Acrylamide/Bis-acrylamide, 30% solution (Sigma-Aldrich, catalog number: A3574)
6× Laemmli SDS sample buffer (Bioland Scientific LLC, catalog number: SAB03-01)
Pierce IP lysis buffer (Thermo Scientific, catalog number: 87787)
Halt protease inhibitor (100×) (Thermo Scientific, catalog number: 87786)
Acetic acid, glacial (Fisher Chemical, catalog number: A38-212)
Glycine 99% (Thermo Scientific, catalog number: A13816.36)
Sodium chloride (NaCl) (Fisher Chemical, catalog number: S271-1)
Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: 208337-1KG)
S.O.C. medium (Invitrogen, catalog number: 15544-034)
Ampicillin sodium salt (Fisher Scientific, catalog number: 69-52-3)
Luria broth base (Miller's LB broth base), powder (Invitrogen, catalog number: 12795027)
LB agar (Lennox L Agar), powder (Invitrogen, catalog number: 22700025)
Solutions
1 M Auxin (IAA) solution in Milli-Q water
50× TAE buffer (see Recipes)
1% agarose gel (see Recipes)
SDS-PAGE 10% resolving protein gel (5 mL)
SDS-PAGE 5% stacking protein gel (see Recipes)
Buffers for SDS-PAGE (see Recipes)
100 mg/mL Ampicillin stock solution
Recipes
50× TAE buffer
Reagent Quantity or Volume Tris-base 242 g Acetic acid 57.1 mL EDTA 100 mL (0.5 M, 8.0 pH) Milli-Q water Adjust volume to 1 L Total 1,000 mL Note: Dilute 50× TAE to 1× TAE (1:50) with Milli-Q water when making agarose gel and running the gel.
1% agarose gel (100 mL)
Reagent Final concentration Quantity or Volume Agarose powder 1% 1 g 1× TAE N/A 100 mL Ethidium bromide 0.5 µg/mL 2–5 µL Total 100 mL In a microwave-safe flask, add 1 g of agarose powder to 100 mL of 1× TAE buffer.
Microwave the flask in short 20–30 s intervals, swirling between each interval. Bring the mixture to a boil until the agarose has completely dissolved and the solution is transparent. Be careful as to not let the solution overboil and evaporate.
Allow mixture to cool before adding 5 µL of ethidium bromide for a 0.5 µg/mL concentration from 10 mg/mL stock. Swirl flask to mix.
Pour solution into casting gel tray and insert well comb. Allow gel to solidify at room temperature (RT).
SDS-PAGE 10% resolving protein gel (5 mL)
Reagent Stock concentration Volume Milli-Q water N/A 1.9 mL Acrylamide mix 30% 1.7 mL Tris-base 1.5 M, 8.8 pH 1.3 mL SDS 10% 50 µL APS 10% 50 µL TEMED N/A 2 µL Total 5 mL SDS-PAGE 5% stacking protein gel (2.0 mL)
Reagent Stock concentration Volume Milli-Q water N/A 1.4 mL Acrylamide mix 30% 0.33 mL Tris-base 1.0 M, 6.8 pH 0.22 mL SDS 10% 20 µL APS 10% 20 µL TEMED N/A 2 µL Total 2 mL Buffers for SDS-PAGE
Reagent Running buffer (1×) Transfer buffer (1×) Glycine 14.4 g 2.9 g Tris-base 3.03 g 5.8 g SDS 1.0 g 0.33 g Milli-Q water 1,000 mL 800 mL Methanol N/A 200 mL Total 1,000 mL 1,000 mL
Laboratory supplies
96-, 24-, 12-, and 6-microwell plates (Thermo Fisher Scientific, catalog number: 130188)
15 mL conical Falcon tube (Fisher Brand, catalog number: 07-200-886)
1.5 mL Eppendorf tubes (Fisher Scientific, catalog number: 02-682-002)
BioLiteTM cell culture treated dishes, 35, 60, and 100 mm Petri dish (Thermo Scientific, catalog numbers: 130180, 130181, and 130182)
WhatmanTM UnifloTM sterile PVDF syringe filters 0.22 µm (Cytiva, catalog number: 9913-2502)
Sterile polystyrene disposable serological pipettes (Fisher Brand, catalog number: 13-678-11E)
Flow cytometry tubes, Mini 35 µm (Olympus Plastics, catalog number: 28-154)
Sterile glass spreader
Equipment
Centrifuge (Eppendorf, model: 5417C and 5804R)
Thermocycler (Eppendorf, model: GX2)
NanoDrop One (Thermo Scientific, model: Nanodrop One Spectrophotoshot Promo)
Flow cytometer (BD-FACS, model: Melody Cell Sorter)
Western blot developer (Azure biosystems, model: 600)
Agarose gel electrophoresis equipment (Bio-Rad, model: Mini-Sub Cell GT)
Protein gel electrophoresis equipment (Bio-Rad, model: Mini PROTEAN Tetra cell)
Spectrophotometer (Vinmax, model: 721-VIS)
Brightfield cell counter (DeNovix, model: CellDrop BF PAYG)
Inverted microscope equipped with a Yokogawa CSU-X1 spinning disc, an Andor iXon Ultra888 EMCCD camera, and a 60× or 100× 1.4 NA Plan-Apochromatic DIC oil immersion objective (Nikon, model: Eclipse TiE)
Software and datasets
Broad Institute sgRNA design tool (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). Alternatively, you can use CHOPCHOP (http://chopchop.cbu.uib.no/) or Crispor (http://crispor.tefor.net/crispor.py)
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Rahi, A., Sodhi, D. K., Magdongon, C. B., Shakya, R. and Varma, D. (2024). Methodology to Create Auxin-Inducible Degron Tagging System to Control Expression of a Target Protein in Mammalian Cell Lines. Bio-protocol 14(2): e4923. DOI: 10.21769/BioProtoc.4923.
- Rahi, A., Chakraborty, M., Agarwal, S., Vosberg, K. M., Agarwal, S., Wang, A. Y., McKenney, R. J. and Varma, D. (2023). The Ndc80-Cdt1-Ska1 complex is a central processive kinetochore–microtubule coupling unit. J. Cell Biol. 222(8): e202208018.
分类
分子生物学 > 蛋白质 > 表达
细胞生物学 > 基于细胞的分析方法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link