- EN - English
- CN - 中文
Detailed Protocol to Perform Direct PCR Using Filamentous Fungal Biomass—Tips and Considerations
使用丝状真菌生物质进行直接 PCR 的详细方案 ——提示和注意事项
发布: 2023年11月05日第13卷第21期 DOI: 10.21769/BioProtoc.4889 浏览次数: 3030
评审: Shweta PanchalXiaofei LiangYueqiang Leng

相关实验方案

用于跨谱应急和现场使用的快速、可持续的热渗透 DNA 提取方案
Stavroula Goudoudaki [...] Yiannis Manoussopoulos
2023年09月05日 1715 阅读
Abstract
The precise and rapid detection of fungi is important in various fields, including clinics, industry, and agriculture. While sequencing universal DNA barcodes remains the standard method for species identification and phylogenetic analysis, a significant bottleneck has been the labor-intensive and time-consuming sample preparation for genomic DNA extraction. To address this, we developed a direct PCR method that bypasses the DNA extraction steps, facilitating efficient target DNA amplification. Instead of extracting genomic DNA from fungal mycelium, our method involves adding a small quantity of mycelium directly to the PCR mixture, followed by a heat shock and vortexing. We found these simple adjustments to be sufficient to lyse many filamentous fungal cells, enabling target DNA amplification. This paper presents a comprehensive protocol for executing direct PCR in filamentous fungi. Beyond species identification, this direct PCR approach holds promise for diverse applications, such as diagnostic PCR for genotype screening without fungal DNA extraction. We anticipate that direct PCR will expedite research on filamentous fungi and diagnosis of fungal diseases.
Key features
• Eliminates the time-consuming genomic DNA extraction step for PCR, enhancing the speed of molecular identification.
• Adds a small quantity of mycelium directly into the PCR mix.
• Emphasizes the crucial role of heat shock and vortexing in achieving efficient target DNA amplification.
• Accelerates the molecular identification of filamentous fungi and rapid diagnosis of fungal diseases.
Graphical overview
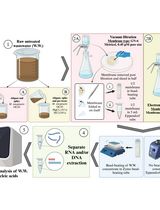
Direct PCR using filamentous fungal biomass
Background
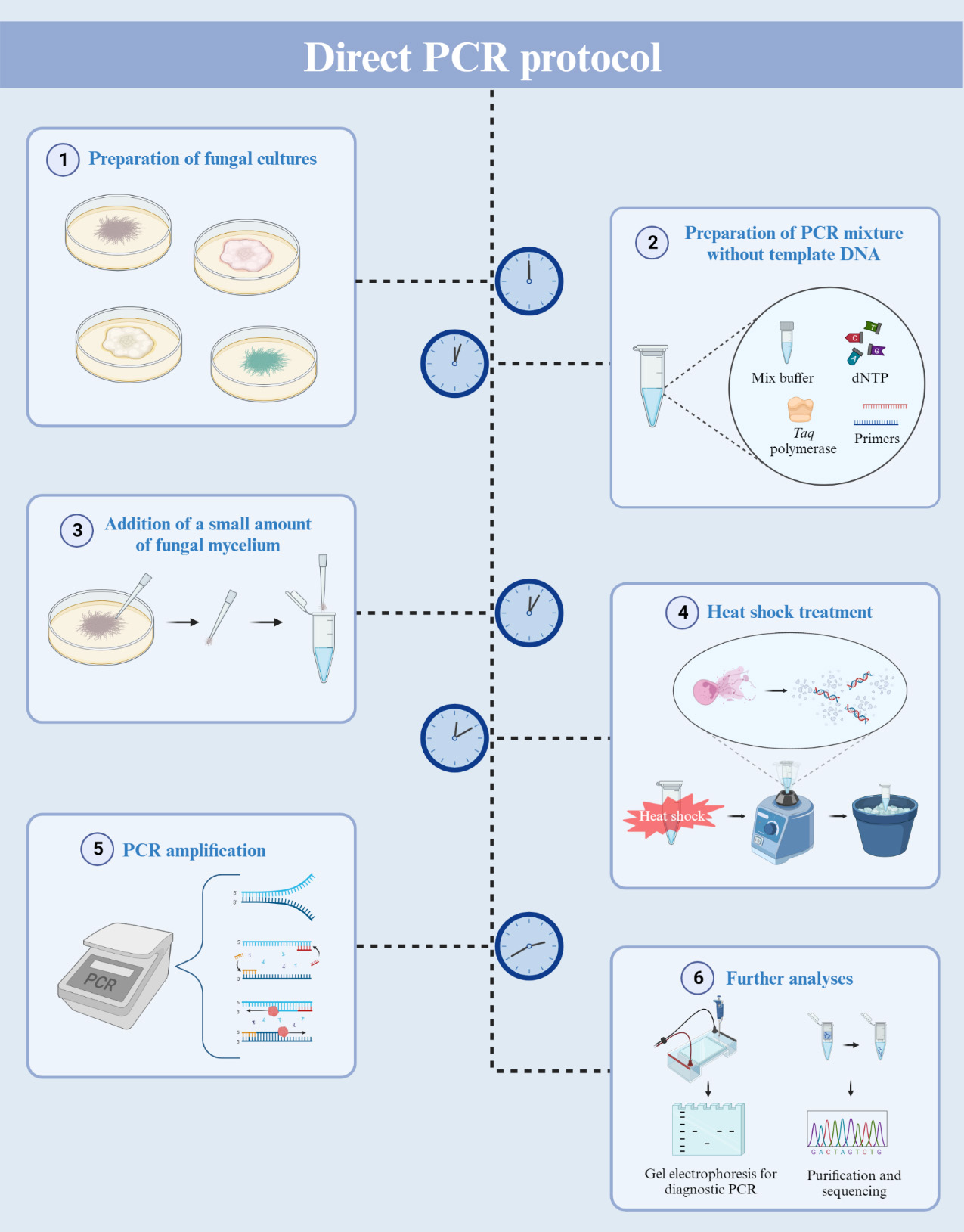
Polymerase chain reaction (PCR) is a powerful tool widely used to amplify specific target regions of DNA. Since its invention in the mid-1980s [1, 2], PCR has become a fundamental technique employed in various fields. A notable application of PCR is in the diagnosis of pathogenic diseases. It allows for the amplification of universal DNA barcodes, facilitating the identification of causative pathogens [3, 4]. Given that multiple copies of DNA are generated from the DNA template, the preparation of this template is essential for performing PCR. For filamentous fungi, the steps for DNA extraction and purification have been prerequisites for PCR amplification. Most fungi possess a rigid cell wall, predominantly constituted of polysaccharides like chitin and glucans [5, 6]. This results in inefficient fungal DNA extraction. The procedure for extracting DNA from fungal sources requires laborious and time-consuming steps involving the grinding of freeze-dried mycelia [7]. Additionally, the fungal DNA extraction process involves multiple stages using toxic chemicals like phenol and chloroform. A technique known as colony PCR involves directly adding cells into the PCR mixture, and it has found widespread application in bacterial and yeast research [8]. In the case of filamentous fungi, previous studies have suggested various PCR methods to minimize fungal DNA extraction steps. The methods proposed by Alshahni et al. and Walch et al. eliminated the need for fungal genomic DNA extraction [9, 10]. However, these methods incorporated additional steps, such as using lysis buffer or bovine serum albumin (BSA), to lyse fungal cell walls. Despite these advancements, the adoption of these methods for filamentous fungi has been limited due to their reduced PCR efficiency and the need for a cell lysis process.
Here, we introduce a direct PCR procedure tailored for filamentous fungi. This approach eliminates the need for the fungal DNA extraction process and the use of additional reagents such as BSA or proteinase. Our method involves adding a small amount of mycelium directly into the PCR mixture, followed by heat shock and vortexing. These heat shock and vortexing steps are key in breaking the fungal cell walls and membrane, releasing the genomic DNA for PCR amplification. This method will expedite the molecular identification of filamentous fungi and facilitate rapid diagnosis of fungal diseases.
Materials and reagents
Potato dextrose broth (Difco, catalog number: 254920)
Agar powder (Duksan, catalog number: 601)
AccuPower® Taq PCR Premix (Bioneer, catalog number: 20-K-2602)
Nuclease-free water (Sigma-Aldrich, catalog number: 7732-18-5)
Primers:
ITS4: 5′-TCCTCCGCTTATTGATATGC-3′
ITS5: 5′- GGAAGTAAAAGTCGTAACAAGG-3′
EF1T: 5′-ATGGGTAAGGAGGACAAGAC-3′
EF2T: 5′-GGAAGTACCAGTGATCATGTT-3′
SeaKem® LE agarose (Lonza, catalog number: 50004)
RedSafe nucleic acid staining solution (iNtRON, catalog number: 21141)
Tris ultrapure (Duchefa Biochemie, catalog number: T1501.1000)
Ethylenediaminetetraacetic acid (EDTA) disodium salt dihydrate (Sigma-Aldrich, catalog number: E5134-50G)
Glacial acetic acid (Sigma-Aldrich, catalog number: PHR1748)
6× Loading buffer (Takara, catalog number: 9156)
100 bp Plus DNA ladder (Bioneer, catalog number: D-1035)
Ethanol EMSURE ACS, ISO, Reag. PH Eur (1 L) (Merck Millipore, catalog number: 1.0098.1011)
Solutions
Culture medium (PDA) (see Recipes)
50× TAE buffer (1 L) (see Recipes)
Recipes
Culture medium (PDA)
Reagent Final concentration Quantity Potato dextrose broth 24 g/L 24 g Agar powder 10 g/L 10 g H2O n/a up to 1 L 50× TAE buffer (1 L)
*Note: Add Tris ultrapure and EDTA disodium salt dihydrate to approximately 700 mL of H2O and stir until dissolved. Carefully add the acetic acid and adjust the volume to 1 L.
Reagent Final concentration Quantity Tris ultrapure 2 M 242 g Glacial acetic acid 1 M 57.1 mL EDTA disodium salt dihydrate 50 mM 18.61 g H2O n/a up to 1 L
Laboratory supplies
Pipette tips (1,000, 200, 10 μL), DNase, RNase, DNA, & endotoxin free (Neptune)
Petri dish (90 mm × 15 mm) (SPL, catalog number 10090)
1.5 mL microtubes (Axygen, catalog number: MCT-15-C)
MEGAquick-spinTM Plus Total Fragment DNA Purification kit (iNtRON, catalog number: 17290)
Equipment
Pipettes (0.5–10 μL, 2–20 μL, 20–200 μL, 100–1,000 μL) (Eppendorf, model: Research Plus®)
Microcentrifuges mini (Labogene, catalog number: LZ-1312)
Vortex Genie 2 (Scientific Industries, catalog number: SI-0256)
MiniAmpTM thermal cycler (Applied Biosystems, catalog number: A37834)
Gel tray L (Takara, catalog number: AD210)
PowerPacTM basic power supply (Bio-Rad, catalog number: 1645050)
Gel imaging system MaXidoc G2 (DAIHAN Scientific, catalog number: DH.WGD00300)
NanoDrop 2000 (Thermo Scientific, catalog number: ND-2000)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Jeon, H., Son, H. and Min, K. (2023). Detailed Protocol to Perform Direct PCR Using Filamentous Fungal Biomass—Tips and Considerations. Bio-protocol 13(21): e4889. DOI: 10.21769/BioProtoc.4889.
分类
微生物学 > 病原体检测 > PCR
分子生物学 > DNA > PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link