- EN - English
- CN - 中文
Identification of Acetylation Sites of Fatty Acid Synthase (FASN) by Mass Spectrometry and FASN Activity Assay
通过质谱法和 FASN 活性测定法鉴定脂肪酸合酶 (FASN) 的乙酰化位点
发布: 2023年11月05日第13卷第21期 DOI: 10.21769/BioProtoc.4873 浏览次数: 1711
评审: Komuraiah MyakalaGautam RunwalAnonymous reviewer(s)

相关实验方案

使用配有超灵敏二极管阵列检测器的高效液相色谱(HPLC)系统从培养细胞和培养基中提取和量化色氨酸和犬尿素
Jeffrey Kim [...] Robert H Weiss
2016年04月05日 9088 阅读
Abstract
Lysine acetylation is a conserved post-translational modification and a key regulatory mechanism for various cellular processes, including metabolic control, epigenetic regulation, and cellular signaling transduction. Recent advances in mass spectrometry (MS) enable the extensive identification of acetylated lysine residues of histone and non-histone proteins. However, protein enrichment before MS analysis may be necessary to improve the detection of low-abundant proteins or proteins that exhibit low acetylation levels. Fatty acid synthase (FASN), an essential enzyme catalyzing the de novo synthesis of fatty acids, has been found to be acetylated in various species, from fruit flies to humans. Here, we describe a step-by-step process of antibody-based protein enrichment and sample preparation for acetylation identification of endogenous FASN protein by MS-based proteomics analysis. Meanwhile, we provide a protocol for nicotinamide adenine dinucleotide phosphate (NADPH) absorbance assay for FASN activity measurement, which is one of the primary functional readouts of de novo lipogenesis.
Key features
• A comprehensive protocol for protein immunoprecipitation and sample preparation for acetylation site identification by mass spectrometry.
• Step-by-step procedures for measurement of FASN activity of fruit fly larvae using an absorbance assay.
Graphical overview
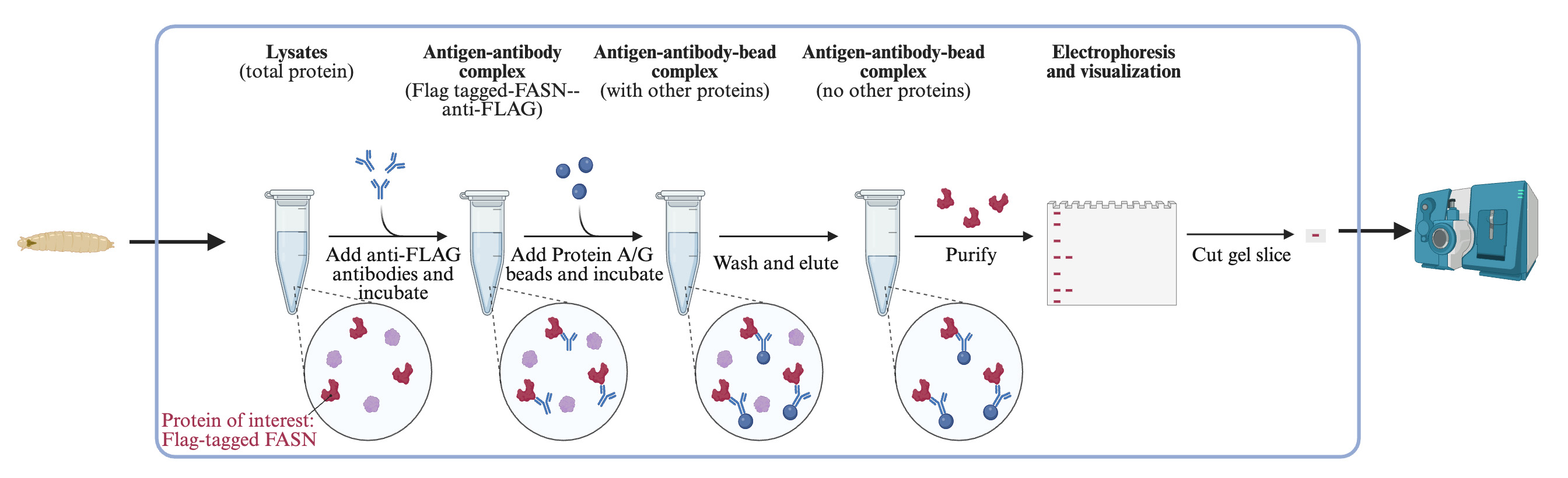
Background
Lysine acetylation is a post-translational modification (PTM) described as the transfer of an acetyl group from acetyl-coenzyme A (acetyl-CoA) to the primary amine of the lysine side chain of a protein. Lysine acetylation links acetyl-CoA metabolism and cellular signaling. Histone acetylation was well known to alter nucleosomal conformation, which is one of the most important regulatory mechanisms of its transcriptional activity (Boffa et al., 1978; Sealy, 1978; Vettese-Dadey et al., 1996). In the past 20 years, thanks to the advances in mass spectrometry (MS)-based proteomics technique, the acetylation of thousands of non-histone proteins was identified and characterized (Choudhary et al., 2009; Wang et al., 2010; Zhao et al., 2010). These proteome-wide discoveries of lysine acetylation sites enable acetylome mapping in various species and in numerous biological processes such as aging and tumor progression, which provides prerequisites for novel discovery of the function of protein acetylation (Sato et al., 2017; Yeo et al., 2020; Zhang et al., 2020). The information on protein acetylation is presented comprehensively in open-access databases such as Protein Lysine Modification Database (PLMD) (Xu et al., 2017).
However, due to the low abundance of some protein targets and relatively low levels of lysine acetylation modification, protein enrichment prior to proteomics is sometimes necessary to improve the efficiency and coverage of the analysis (Jensen et al., 2021). An antibody-based enrichment method is one of the most widely used strategies for the characterization of the dynamic acetylome or modifications of a protein of interest (Inuzuka et al., 2012; Lin et al., 2013; Miao et al., 2022). Here, we describe step-by-step procedures for the identification of acetylated lysine sites of fatty acid synthase (FASN) of fruit fly Drosophila larvae.
De novo lipogenesis (DNL) is a tightly regulated metabolic pathway that converts carbohydrates to fatty acids (Song et al., 2018). FASN uses acetyl-CoA and malonyl-CoA as the substrates and nicotinamide adenine dinucleotide phosphate (NADPH) as a cofactor to catalyze the formation of palmitate, the first fatty acid product of DNL. Therefore, FASN activity is one of the primary functional readouts for DNL. Here, we describe an NADPH absorbance assay that can be used to quantify FASN enzymatic activity and kinetics (Dils and Carey, 1975; Cox and Hammes, 1983; Menendez et al., 2004).
Materials and reagents
Flies
Fly lines used in this study: ywR (a gift from Eric Rulifson); ywR; FASNFlag (Miao et al., 2022). Flies were maintained at 25 °C, 60% relative humidity, and a 12:12 h light/dark cycle. Adults and larvae are reared on standard cornmeal and yeast-based food.
Reagents
PierceTM IP lysis buffer (Thermo ScientificTM, catalog number: 87787)
Protease inhibitor cocktail (PIC, 100×) (Sigma-Aldrich, catalog number: P8340)
PMSF protease inhibitor (Fisher Scientific, Thermo ScientificTM, catalog number: PI36978)
Sodium butyrate (VWR, Thermo ScientificTM, catalog number: AAAA11079-06)
Nicotinamide (NAM) (Sigma-Aldrich, catalog number: 72340)
PierceTM BCA Protein Assay kit (Thermo ScientificTM, catalog number: 23225)
SureBeadsTM Protein G magnetic beads (Bio-Rad, catalog number: 1614023)
2× Laemmli sample buffer (Bio-Rad, catalog number: 1610737)
10× Premixed electrophoresis buffer (Bio-Rad, catalog number: 1610732)
4%–20% Mini-PROTEAN® TGXTM Precast protein gels (Bio-Rad, catalog number: 4561093)
Sucrose (Sigma-Aldrich, catalog number: S9378)
GibcoTM HEPES (1 M) (Fisher Scientific, catalog number: 15-630-080)
Magnesium chloride (MgCl2) (Fisher Scientific, Thermo Scientific Chemicals, catalog number: AC223210010)
Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632)
UltraPureTM 0.5 M EDTA, pH 8.0 (Fisher Scientific, InvitrogenTM, catalog number: 15-575-020)
PierceTM Coomassie (Bradford) Protein Assay kit (Thermo ScientificTM, catalog number: 23200)
Monoclonal ANTI-FLAG® M2 antibody (Sigma-Aldrich, catalog number: F1804-50UG)
Solutions
100 mM PMSF
5 M Sodium Butyrate
1 M NAM
1 M Sucrose
1 M MgCl2
1 M DTT
1 M Dipotassium phosphate (K2HPO4)
1 M Potassium dihydrogen phosphate (KH2PO4)
24 mM NADPH
6.2 mM Acetyl-CoA
5.8 mM Malonyl-CoA
Protein lysis buffer for immunoprecipitation and electrophoresis (lysis buffer 1) (see Recipes)
Protein denature buffer for protein elution and electrophoresis (denature buffer) (see Recipes)
Electrophoresis running buffer (see Recipes)
Protein lysis buffer for FASN activity assay (lysis buffer 2) (see Recipes)
200 mM potassium phosphate buffer (PPB), pH 6.6 (see Recipes)
FASN reaction buffer (see Recipes)
Recipes
Protein lysis buffer for immunoprecipitation and electrophoresis (lysis buffer 1)
6–7 mL of lysis buffer 1 will be used for lysis and washing steps; calculate the total volume according to number of samples and make the buffer freshly.
PierceTM IP lysis buffer supplied with 1× PIC, 1 mM PMSF, 50 mM sodium butyrate, and 50 mM NAM
Protein denature buffer for protein elution and electrophoresis (denature buffer)
Note: Make fresh.
950 μL of 2× Laemmli sample buffer
50 μL of BME
1 mL of lysis buffer 1
Electrophoresis running buffer
Note: Can be stored at room temperature.
10× Premixed electrophoresis buffer following dilution to 1× with water
Protein lysis buffer for FASN activity assay (lysis buffer 2)
300 μL of lysis buffer 2 will be used for the lysis step; calculate the total volume according to number of samples and make the buffer freshly.
250 mM sucrose
20 mM HEPES Buffer, pH 7.2
2 mM MgCl2
1 mM DTT
1 mM EDTA
1× PIC
200 mM potassium phosphate buffer (PPB), pH 6.6
Note: Stable at 4 °C for two months.
0.76 mL of 1 M K2HPO4
1.24 mL of 1 M KH2PO4
Bring volume to 10 mL with distilled water
FASN reaction buffer
200 μL of the reaction buffer will be added to each sample/standard; calculate the volume according to number of standards and samples and make the buffer freshly.
200 mM PPB, pH 6.6
1 mM DTT
1 mM EDTA
0.24 mM NADPH
0.031 mM Acetyl-CoA
Laboratory supplies
Pellet PestleTM cordless motor (Fisher Scientific, catalog number: 12-141-361)
Magnetic bead rack (Bio-Rad, catalog number: 1614916)
1.7 mL low-binding microcentrifuge tube (Fisher Scientific, catalog number: 07-200-184)
Equipment
Sonifier (Branson, model: SFX150)
Microcentrifuge (SorvallTM LegendTM Micro 21)
Digital dry baths/block heaters (Fisher Scientific, catalog number: 88860103)
Microplate spectrophotometer (BioTek Epoch 2)
ChemiDoc MP Imaging System (Bio-Rad)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Miao, T. and Bai, H. (2023). Identification of Acetylation Sites of Fatty Acid Synthase (FASN) by Mass Spectrometry and FASN Activity Assay. Bio-protocol 13(21): e4873. DOI: 10.21769/BioProtoc.4873.
分类
细胞生物学 > 细胞新陈代谢 > 氨基酸
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










