- EN - English
- CN - 中文
Spot Assay and Colony Forming Unit (CFU) Analyses–based sensitivity test for Candida albicans and Saccharomyces cerevisiae
基于斑点分析和菌落形成单位(CFU)分析的白色念珠菌和酿酒酵母敏感性测试
发布: 2023年11月05日第13卷第21期 DOI: 10.21769/BioProtoc.4872 浏览次数: 2750
评审: Alba BlesaFernando A Gonzales-ZubiateThibaud T. RenaultAnonymous reviewer(s)
Abstract
Cellular sensitivity is an approach to inhibit the growth of certain cells in response to any non-permissible conditions, as the presence of a cytotoxic agent or due to changes in growth parameters such as temperature, salt, or media components. Sensitivity tests are easy and informative assays to get insight into essential gene functions in various cellular processes. For example, cells having any functionally defective genes involved in DNA replication exhibit sensitivity to non-permissive temperatures and to chemical agents that block DNA replication fork movement. Here, we describe a sensitivity test for multiple strains of Saccharomyces cerevisiae and Candida albicans of diverged genetic backgrounds subjected to several genotoxic chemicals simultaneously. We demonstrate it by testing the sensitivity of DNA polymerase defective yeast mutants by using spot analysis combined with colony forming unit (CFU) efficiency estimation. The method is very simple and inexpensive, does not require any sophisticated equipment, can be completed in 2–3 days, and provides both qualitative and quantitative data. We also recommend the use of this reliable methodology for assaying the sensitivity of these and other fungal species to antifungal drugs and xenobiotic factors.
Keywords: Spot assays (斑点测定)Background
Sensitivity assays are crucial in determining the susceptibility of a cell to a particular stress and are used to gain insights into diverse cellular processes in both prokaryotic and eukaryotic cells, including yeasts like Saccharomyces cerevisiae, Candida albicans, or Schizosaccharomyces pombe (Kwolek-Mirek and Zadrag-Tecza, 2014). S. cerevisiae and C. albicans are genetically related fungal species and are considered the best model organisms for sensitivity studies in yeast research due to their ease of manipulation, rapid growth, and well-established genetic modification strategies (Botstein et al., 1997; Berman, 2012). The growth curve analysis based on the optical density (OD) measurement in the presence and absence of a cytotoxic chemical is routinely used to monitor the sensitivity of a strain. Here, we describe an alternative protocol that uses a combination of spot and CFU analyses to obtain both qualitative and quantitative sensitivity results of several strains in a single experiment in a short time. We have been using this methodology repeatedly and successfully in our studies for many years now (Manohar et al., 2018; Kumari et al., 2021 and 2023; Manohar et al., 2022; Patel et al., 2023).
In the spot assay, yeast cultures are spotted on agar plates with or without different concentrations of any cytotoxic agents in a dilution series; sensitivity is analyzed based on the density of the cells present in a given spot. Comparison of growth inhibition in a specific spot with and without drugs allows to assess the sensitivity of a strain to that particular drug. Similarly, relative sensitivity to a particular drug can be assessed by comparing various isogenic wildtype and mutant strains. Since spotting will only determine the cumulative qualitative behavior of a group of colonies present in a particular spot, we support this assay by enumerating colony forming unit (CFU) efficiency of a yeast strain by spreading different dilutions of the culture on solid media plates with or without different concentrations of drugs. Estimating the number of colonies and their comparison across drug concentrations or strains makes it a quantifiable sensitivity test. The combination of both assays provides us with a more comprehensive sensitivity profile of a strain. The complete assay involves spotting and spreading serially diluted yeast cells of culture on agar plates; the resulting growth patterns in both types of plates are then used to assess the sensitivity of each strain (Figure 1).
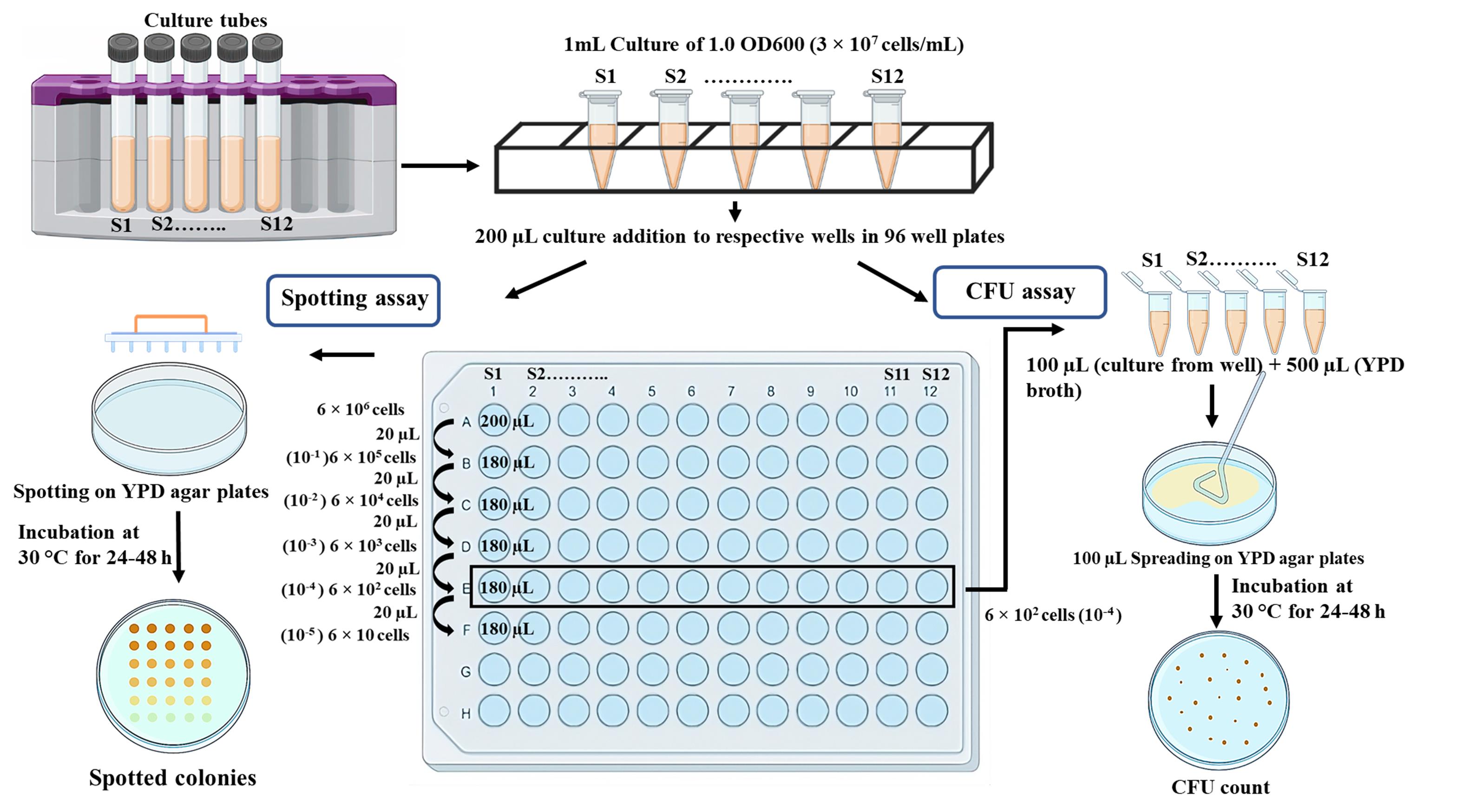
Figure 1. Schematic representation of the methodology to determine sensitivity of isogenic strains to a specific drug or reagent
This protocol shows the steps involved in testing four strains in their sensitivity to the presence of hydroxyurea (HU), a DNA replication inhibitor, and methyl methanesulfonate (MMS), a DNA methylating agent. However, the sensitivity of 12 different strains or drugs can be verified using a 96-well plate. We used wildtype strains of S. cerevisiae and C. albicans and their pol32 null strains for sensitivity tests. Pol32 is the smallest subunit of DNA polymerase delta, a DNA polymerase involved in both lagging and leading strand synthesis (Acharya et al., 2011; Khandagale et al., 2019). In its absence, cells exhibit high temperature sensitivity and growth retardation in the presence of HU, MMS, and other DNA damaging agents (Patel et al., 2023). Here, we repeated the experiment to show the effect of HU and MMS on the growth of these strains by simultaneously using spot and CFU analyses (Figure 2). The differential cell density among the strains in the spot assay and the statistical estimation of the number of colonies in the CFU assay confirmed that pol32-deficient strains are more sensitive to these tested drugs than their respective wildtype strains.
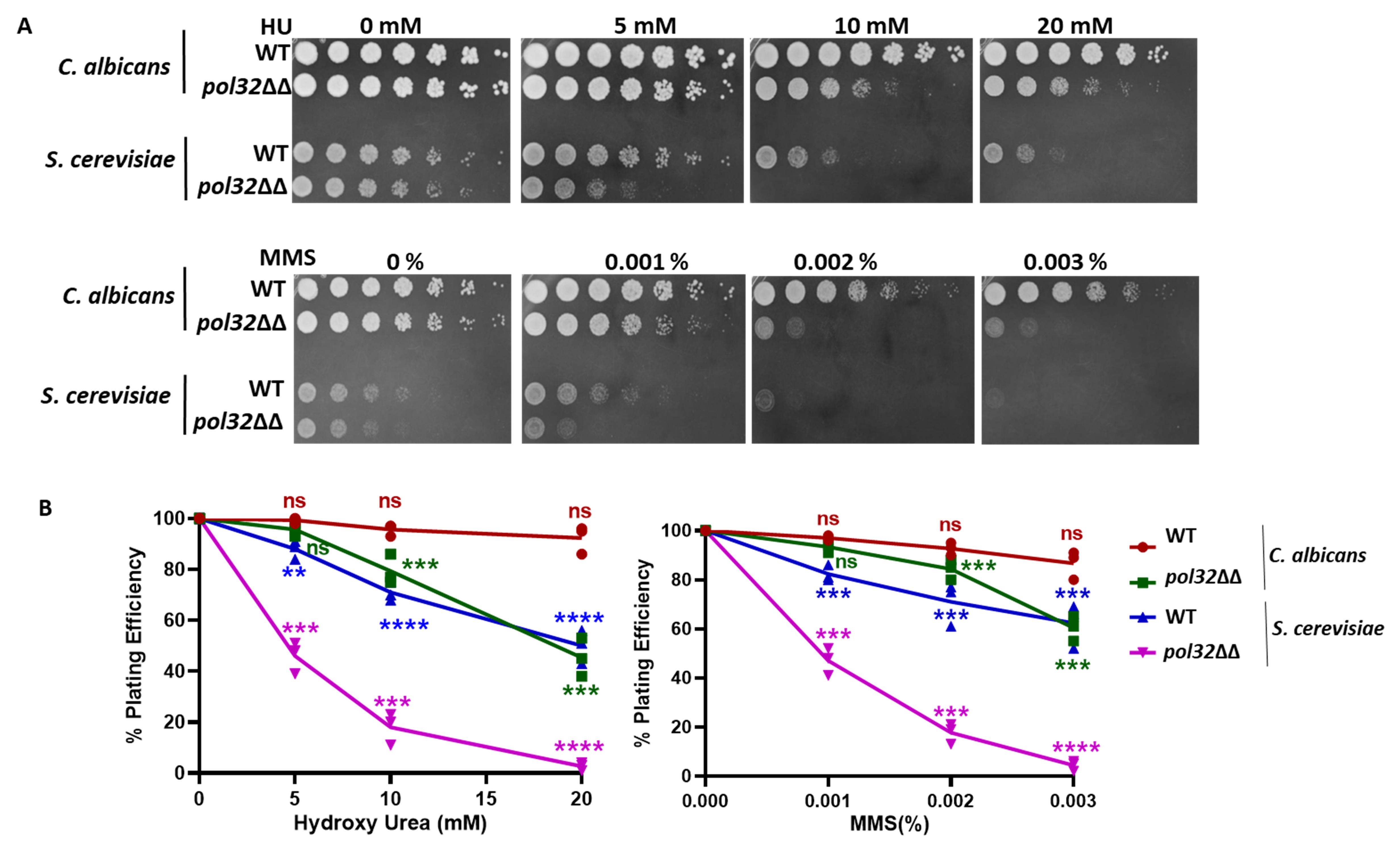
Figure 2. Sensitivity tests for pol32 deficient strains of C. albicans and S. cerevisiae. (A) Cells of wildtype (WT) and pol32 knockout strains of C. albicans (SC5314, diploid) and S. cerevisiae (EMY74.7, haploid) were spotted on YPD-agar media containing different concentrations of hydroxyurea (HU) and methyl methanesulfonate (MMS). (B) Dilutions of these cells were spread to get isolated colonies; colonies were counted and plotted in a line graph as obtained from Table 1 and Table S1. ns = nonsignificant and *** p value < 0.001.
This protocol is simple, cost effective, and does not require any expensive equipment. This assay can be used to assess multiple parameters such as growth phenotypes, viability, stress or drug resistance, and genotoxicity of multiple strains at a time (Kumari et al., 2023). Altogether, this procedure provides a well-standardized, sensitive, and reproducible protocol to assess yeasts’ sensitivity.
Materials and reagents
Yeast strain of interest
Round-bottom glass culture tubes (Borosil, catalog number: 9900006)
250 mL conical flask (Schott Duran, catalog number: 1006940)
Yeast extract peptone dextrose (YPD) (Himedia, catalog number: M1363)
Yeast extract peptone dextrose agar (YPDA) (Himedia, catalog number: G038)
Ethanol (100% and 70%) (Fisher Chemical, catalog number: 2051537)
Hydroxyurea (MP Biomedical, catalog number: 102023)
MMS (SRL, catalog number: 74384)
Sterile 1.5 mL microcentrifuge tube (Axygen, catalog number: MCT150LC)
Round U-bottom 96-well plate (Thermo Fisher Scientific, catalog number: 165306)
Sterile Petri dish 100 mm × 15 mm (Falcon, catalog number: 351008)
Glass Petri dish 100 mm × 15 mm (Borosil, catalog number: 3165077)
Sterile spreader (Tarson, catalog number: 920081)
Cuvettes 200–1,600 nm (Eppendorf, catalog number: 0030106300)
Aluminum foil (Rolias)
Gloves (Blue Shield)
Equipment
Laminar airflow (Thermo Scientific Biological Safety Cabinets, catalog number: 41346502)
Sterile pipette sets (Rainin, catalog number: E1338890T-SL1000, B637036181-SL200, B641141423-SL20)
Laboratory spirit lamp or Bunsen burner (VWR, catalog number: 17822-605)
Spectrophotometer (Eppendorf, Bio Photometer Plus, catalog number: 6132)
48-pin spotter (Sigma-Aldrich, catalog number: R2383)
Refrigerated shaker incubator (Scigenic Biotech, catalog number: LE-4676-AH)
Refrigerated incubator (Scigenic Biotech, catalog number: C-1NC-100)
Chemidoc XRS gel imager (Bio-Rad, catalog number: 1708370)
White Light box (Cole-Parmer: NC1851470)
Software
Bio-Rad Image lab (2017, version number: 6.0.1)
GraphPad prism v8.0
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Sahu, S. R., Utkalaja, B. G., Patel, S. K. and Acharya, N. (2023). Spot Assay and Colony Forming Unit (CFU) Analyses–based sensitivity test for Candida albicans and Saccharomyces cerevisiae. Bio-protocol 13(21): e4872. DOI: 10.21769/BioProtoc.4872.
分类
微生物学 > 微生物遗传学
微生物学 > 微生物细胞生物学 > 细胞活力
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link














