- EN - English
- CN - 中文
Purification of Long Non-coding RNAs on Replication Forks Using iROND (Isolate RNAs on Nascent DNA)
使用 iROND 纯化复制叉上的长非编码 RNA(分离新生 DNA 上的 RNA)
发布: 2023年11月05日第13卷第21期 DOI: 10.21769/BioProtoc.4869 浏览次数: 2078
评审: Emilia KrypotouThirupugal GovindarajanZheng Zachory WeiDr. Amit K. Tripathi

相关实验方案

基于适体的 mRNA 亲和纯化程序 (RaPID) 用于鉴定酵母中的相关 RNA (RaPID-seq) 和蛋白质 (RaPID-MS)
Rohini R. Nair [...] Jeffrey E. Gerst
2022年01月05日 5399 阅读
Abstract
Fork stability is key to genome DNA duplication and genetic integrity. Long non-coding RNAs (LncRNAs) may play vital roles in fork stabilization and chromatin remodeling. Existing techniques such as NCC-RNA sequencing are useful to identify LncRNAs on nascent chromatin DNA. However, there is still a lack of methods for LncRNAs purification directly from replicative forks, hindering a deep understanding of the functions of LncRNAs in fork regulation. Here, we provide a step-by-step protocol named iROND (isolate RNAs on nascent DNA). iROND was developed and modified from iPOND, a well-known method for purifying fork-associated proteins. iROND relies on click chemistry reaction of 5'-ethynyl-2'-deoxyuridine (EdU)-labeled forks and biotin. After streptavidin pull down, fork-associated LncRNAs and proteins are purified simultaneously. iROND is compatible with downstream RNA sequencing, qPCR confirmation, and immunoblotting. Integrated with functional methods such as RNA fluorescent in situ hybridization (RNA FISH) and DNA fiber assay, it is feasible to screen fork-binding LncRNAs in defined cell lines and explore their functions. In summary, we provide a purification pipeline of fork-associated LncRNAs. iROND is also useful for studying other types of fork-associated non-coding RNAs.
Key features
• Purify long non-coding RNAs (LncRNAs) directly from replication forks.
• Connects to RNA sequencing for screening easily.
• Allows testing various genotoxic stress responses.
• Provides LncRNA candidate list for downstream functional research.
Graphical overview
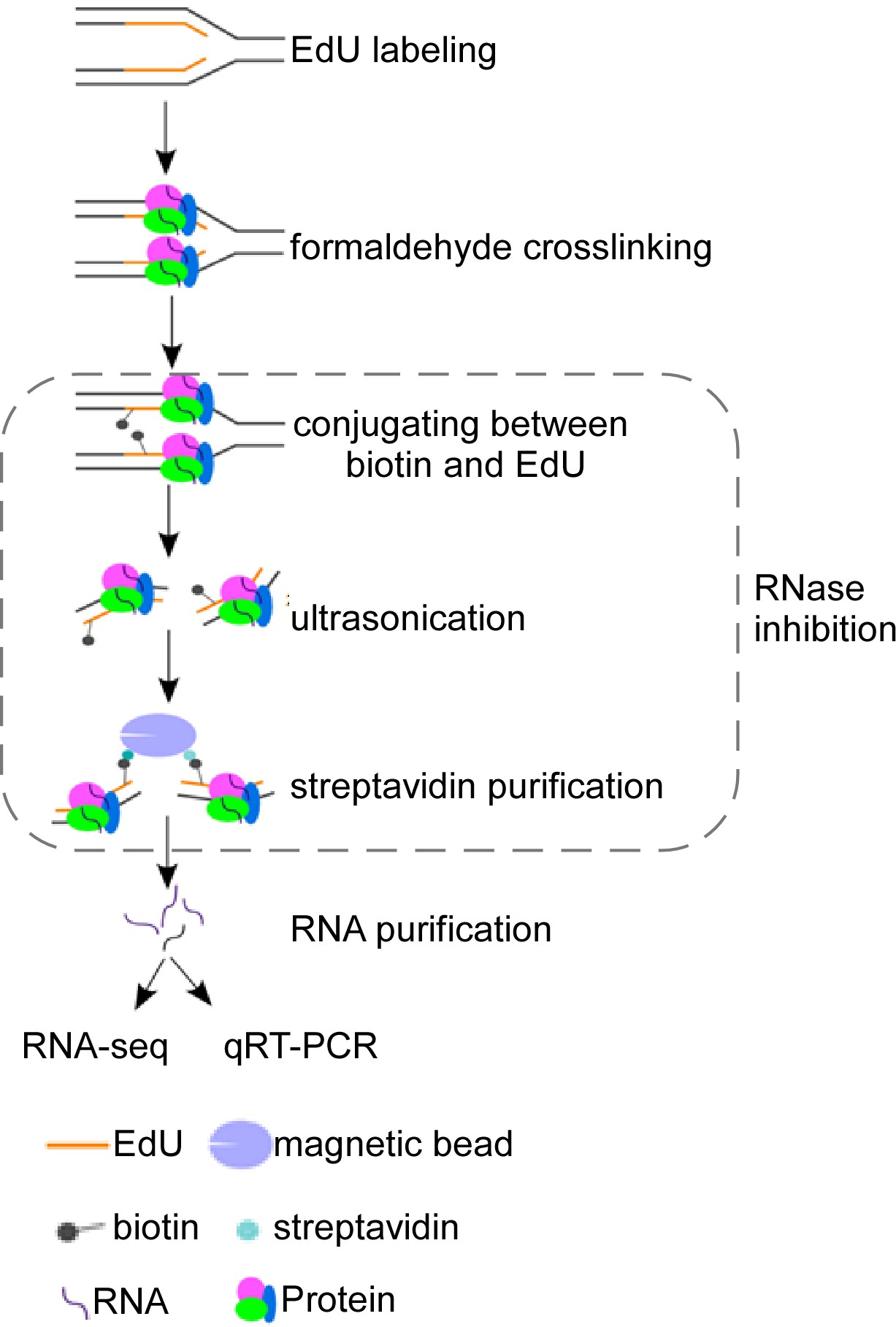
Schematic overview of isolate RNAs on nascent DNA (iROND) protocol. Cells were pulse-labeled with 5'-ethynyl-2'-deoxyuridine (EdU) for 10 min before paraformaldehyde fixation. EdU-positive forks were ligated with biotin through Click-IT chemistry reaction. Genomic DNA was ultrasonically cracked and crosslinked with streptavidin for pulling down. Both RNA and protein components were purified. RNA components were used for downstream RNA sequencing and qPCR validation. Protein components were used for immunoblotting to evaluate binding dynamics of fork-associated proteins such as helicase, topoisomerase, and DNA polymerases.
Background
DNA replication and DNA repair are central topics in genome stability research. Currently, a vast majority of studies focuses on protein components intensively. Through the investigation of core factors such as ATM, ATR, CHK1, CHK2, DNA-PK, and P53, as well as their associated signaling pathways, the fundamental regulatory mechanisms in DNA replication and DNA repair have been revealed (Jackson and Bartek, 2009). For example, using isolate proteins on nascent DNA (iPOND) technology, numerous replication fork-associated protein components have been identified, including DNA polymerases, helicases, and epigenetic modifiers (Sirbu et al., 2012). The initiation, elongation, and termination of steady-state replication forks are strictly controlled to ensure the integrity of the genome replication process (Dungrawala and Cortez, 2015). In order to cope with genotoxic drug-induced fork collapse or stalling, endogenous DNA replication stress response mechanisms can ensure DNA replication progression and reduce the accumulation of DNA breaks (Berti et al., 2020). In terms of DNA repair, numerous specific techniques have been developed. For instance, GFP-based reporter systems were used to evaluate DNA repairing efficiency (Pierce et al., 1999). Single-cell comet assay is used broadly to examine general DNA damage levels (Collins, 2004). Overall, the involvement of protein components in genome stability maintenance is presently well understood.
Besides protein components, a few recent studies raised the potential importance of RNA components in DNA repair and replication, facilitated by indirect identification techniques. For example, the NCC-RNA-seq technique, based on the principle of biotin affinity purification, has been used to identify multiple long and short RNA molecules associated with nascent chromosomes (Gylling et al., 2020). The functions of these RNAs in chromosome reshaping and maturation warrant further investigation. Additionally, using RNA affinity purification coupled with mass spectrometry, the long non-coding RNA (LncRNA) NORAD plays a critical role in genome stability through interaction with PUMILIO protein (Elguindy and Mendell, 2021). Comparative transcriptomics is an effective approach for discovering functional non-coding RNAs. In our previous studies, we identified a novel LncRNA, DISCN, by comparing the expression profiles of LncRNAs in mouse embryonic stem cells (mESCs) with differentiated cells. DISCN exhibits specific high expression in embryonic and neural stem cells. DISCN forms a functional complex with the nucleolar protein nucleolin and the DNA single-strand binding protein RPA, through which it keeps an RPA protein pool to ensure the efficiency of DNA replication stress response and DNA repair (Wang et al., 2021). These studies support the notion that non-coding RNAs form a new regulatory layer in genomic stability system. However, limited by direct purification techniques, the roles of non-coding RNAs in DNA replication or DNA repair are still not understood.
We have recently developed a novel method called iROND (isolate RNAs on nascent DNA). Using iROND, we identified a specific fork-binding LncRNA Lnc956, in the embryonic stem cell system. Lnc956 forms a stable nucleic acid–protein complex with TRIM28 and HSP90B1, facilitating the integration of the CMG helicase at replication fork sites and ensuring replication fork stability in mESCs (Zhang et al., 2023). Here, we provide a step-by-step protocol for iROND. iROND is applicable in different types of rapidly proliferating cell lines and compatible with downstream functional validation systems such as RNA fluorescence in situ hybridization (RNA-FISH) and DNA fiber assays.
Materials and reagents
Biological materials
Mouse embryonic stem cells (derived from C57Bl/6J blastocysts)
Mouse NIH3T3 cells (kindly provided by Dr. Bingyu Mao at Kunming Institute of Zoology, Chinese Academy of Sciences)
Materials
35 mm cell culture dish (Corning, catalog number: 430165)
60 mm cell culture dish (NEST, catalog number: 705001)
100 mm culture dish (NEST, catalog number: 704001)
150 mm culture dish (Corning, catalog number: 430599)
200 μL PCR tube (Axygen, catalog number: AXYPCR02LC)
1.5 mL centrifugal tube (Axygen, catalog number: AXYMCT150C)
15 mL centrifugal tube (NEST, catalog number: 601052)
50 mL centrifugal tube (NEST, catalog number: 602002)
0.22 μm filter (Millipore, catalog number: SLGP033RB)
Reagents
5-ethynyl-2′-deoxyuridine (EdU) (Life Technology, catalog number: A10044)
Thymidine (Sigma-Aldrich, catalog number: T1895)
37% (w/v) formaldehyde solution (Sigma-Aldrich, catalog number: F1635); caution: toxic
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888)
Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P3911)
Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655)
Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S5136)
Glycine (Sigma-Aldrich, catalog number: G7126)
Triton X-100 (Sigma-Aldrich, catalog number: X100)
BSA (Sigma-Aldrich, catalog number: V900933)
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418)
Copper (II) sulfate pentahydrate (CuSO4·5H2O) (Sigma-Aldrich, catalog number: 209198)
(+) Sodium L-ascorbate (Sigma-Aldrich, catalog number: A4034)
Biotin azide (Invitrogen, catalog number: B10184)
SDS (Sangon Biotech, catalog number: A600485)
Tris (Sangon Biotech, catalog number: A610195)
Glycerol (Sangon Biotech, catalog number: A600232)
Bromophenol blue (Sangon Biotech, catalog number: A602230)
EDTA (Sigma-Aldrich, catalog number: E9884)
Agarose (Merck, catalog number: A4718)
Dithiothreitol (DTT) (Merck, catalog number: D8255)
Aprotinin (Merck, catalog number: A6279)
Leupeptin (Merck, catalog number: L2884)
Streptavidin agarose (Novagen, catalog number: 69203-3)
RNase inhibitor (Thermo Fisher, catalog number: EO0381)
Diethyl pyrocarbonate (DEPC) (Sigma-Aldrich, catalog number: D5758)
Protease K (Thermo Fisher, catalog number: 4333793)
Chloroform (Sigma-Aldrich, catalog number: 366911)
Isopropyl alcohol (Sigma-Aldrich, catalog number: W292912)
Ammonium acetate (Sigma-Aldrich, catalog number: A7262)
Alcohol (Sigma-Aldrich, catalog number: AX0442)
Acetic acid (Sigma-Aldrich, catalog number: 695092)
TRIzol (Thermo Fisher, catalog number: 15596026)
Hydroxyurea (HU) (Sigma-Aldrich, catalog number: H8627)
EdU solution (see Recipes)
DEPC-H2O (see Recipes)
Thymidine solution (see Recipes)
Chase medium (see Recipes)
1× PBS solution (see Recipes)
1% Formaldehyde solution (see Recipes)
1.25 M Glycine solution (see Recipes)
Permeabilization buffer (see Recipes)
Wash buffer (see Recipes)
Biotin-azide solution (see Recipes)
100 mM CuSO4 solution (see Recipes)
Sodium L-ascorbate solution (see Recipes)
Lysis buffer (pH 8.0) (see Recipes)
Salt wash buffer (see Recipes)
2× SDS Laemmli sample buffer (2× SB) (see Recipes)
50× Tris acetate-EDTA (TAE) buffer (see Recipes)
EdU solution
Reagent (use for storing) Final concentration Quantity EdU (50 mg) 10 mM 25.2 mg DMSO n/a 10 mL Total n/a 10 mL Reagent (use for cell labeling) Final concentration Quantity EdU (10 mM) 10 μM 1 μL mESCs medium (see Step A1) n/a 999 μL Total n/a 1 mL DEPC-H2O
Reagent Final concentration Quantity DEPC n/a 1 mL H2O n/a 999 mL Total n/a 1,000 mL Thymidine solution
Reagent (use for storing) Final concentration Quantity Thymidine (1 g) 10 mM 24.2 mg DEPC-H2O n/a 10 mL Total n/a 10 mL Chase medium
Reagent (use for cell labeling) Final concentration Quantity Thymidine (10 mM) 10 μM 1 μL mESCs Medium n/a 999 μL Total n/a 1 mL 1× PBS solution
Reagent Final concentration Quantity NaCl 137 mM 8 g KCl 3 mM 0.2 g Na2HPO4 8 mM 1.15 g KH2PO4 2 mM 0.24 g DEPC-H2O n/a 1 L Total n/a 1 L 1% Formaldehyde solution
Reagent Final concentration Quantity Formaldehyde (37%) 1% 0.27 mL DEPC-PBS n/a 9.73 mL Total n/a 10 mL 1.25 M Glycine solution
Reagen Final concentration Quantity Glycine 1.25 M 46.92 g DEPC-H2O n/a 500 mL Total n/a 500 mL Permeabilization buffer
Reagent Final concentration Quantity Triton X-100 0.25% 2.5 mL DEPC-PBS n/a 997.5 mL Total n/a 1 L Wash buffer
Reagent Final concentration Quantity BSA 0.5% 0.5 g DEPC-PBS n/a 100 mL Total n/a 100 mL Biotin-azide solution
Reagent Final concentration Quantity Biotin-azide 1 mM 1 mg DMSO n/a 1.624 mL Total n/a 1.624 mL 100 mM CuSO4 solution
Reagent Final concentration Quantity CuSO4·5H2O 100 mM 1.248 g DEPC-H2O n/a 50 mL Total n/a 50 mL Sodium L-ascorbate solution
Reagent Final concentration Quantity (+) sodium L-ascorbate 100 mM 20 mg DEPC-H2O n/a 1 mL Total n/a 1 mL Lysis buffer (pH 8.0)
Reagen Final concentration Quantity Tris 10 mM 1.21 g SDS 7 mM 2 g DEPC-H2O n/a 200 mL RNase inhibitor 10 U/μL 200 μg Aprotinin 1 μg/mL 200 μg Leupeptin 1 μg/mL 200 μg Total n/a 200 mL Salt wash buffer
Reagent Final concentration Quantity NaCl 1 M 14.625 g DEPC-H2O n/a n/a Total n/a 250 mL 2× SDS Laemmli sample buffer (2× SB)
Reagent Final concentration Quantity SDS 123 mM 0.4 g Bromophenol blue 1 mM 0.01 g Glycerol 18% 2 mL 1 M Tris (pH 6.8)
1M DTT
1 mM
1 mM
1.25 mL
1.25 mL
H2O n/a 8 mL Total n/a 12.5 mL 50× Tris acetate-EDTA (TAE) buffer
Reagent Final concentration Quantity Tris 2 M 242 g Acetic acid n/a 57.1 mL EDTA 0.1 M 37.2 g H2O n/a 942.9 mL Total n/a 1,000 mL
Equipment
Thermo Forma series II water jacketed CO2 incubator (Thermo Scientific, model: 3111)
Nikon eclipse Ti inverted microscope (Nikon, model: Ti)
Ultrasonic cell disruptor (Diagenode, Bioruptor Plus)
90 µm nylon mesh (Small Parts Inc., catalog number: B000FN0PGQ)
Roll shaker for incubation (Thermo Scientific, model: PHMT)
Real-time fluorescence quantitative PCR instrument (Bio-Rad, model: CFX96Touch)
Vortexer (VWR analog vortex mixer, catalog number: 10153-838)
4 °C refrigerator (Haier, model: HYC-1090)
-20 °C freezer (Haier, model: DWL262)
NanoDrop (Thermo Scientific, ND2000)
-80 °C freezer (Thermo Scientific, model: TSX60086VRAKLB)
Software
Microsoft Excel (Microsoft, https://products.office.com/en-us/excel) (Office Excel 2016, September 22, 2015)
GraphPad Prism (GraphPad, https://www.graphpad.com/scientific-software/prism/) (GraphPad Prism 8.3.0, October 29, 2019)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zhang, W., Tang, M., Wang, L., Zheng, P. and Zhao, B. (2023). Purification of Long Non-coding RNAs on Replication Forks Using iROND (Isolate RNAs on Nascent DNA). Bio-protocol 13(21): e4869. DOI: 10.21769/BioProtoc.4869.
- Zhang, W., Tang, M., Wang, L., Zhou, H., Gao, J., Chen, Z., Zhao, B. and Zheng, P. (2023). Lnc956 -TRIM28-HSP90B1 complex on replication forks promotes CMG helicase retention to ensure stem cell genomic stability and embryogenesis. Sci. Adv. 9(4): eadf6277.
分类
分子生物学 > RNA > RNA 纯化 > 亲和纯化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











