- EN - English
- CN - 中文
Efficient Large DNA Fragment Knock-in by Long dsDNA with 3′-Overhangs Mediated CRISPR Knock-in (LOCK) in Mammalian Cells
在哺乳动物细胞中通过具有 3' 突出端的长 dsDNA 介导的 CRISPR 敲入 (LOCK) 实现高效的大 DNA 片段敲入
发布: 2023年10月20日第13卷第20期 DOI: 10.21769/BioProtoc.4853 浏览次数: 2786
评审: Xin XuAnonymous reviewer(s)

相关实验方案

利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1253 阅读
Abstract
An efficient and precise genome-editing approach is in high demand in any molecular biology or cell biology laboratory worldwide. However, despite a recent rapid progress in the toolbox tailored for precise genome-editing, including the base editors and prime editors, there is still a need for a cost-effective knock-in (KI) approach amenable for long donor DNA cargos with high efficiency. By harnessing the high-efficient double-strand break (DSB) repair pathway of microhomology-mediated end joining, we previously showed that a specially designed 3′-overhang double-strand DNA (odsDNA) donor harboring 50-nt homology arm (HA) allows high-efficient exogenous DNA KI when combined with CRISPR-Cas9 technology. The lengths of the 3′-overhangs of odsDNA donors could be manipulated by the five consecutive phosphorothioate (PT) modifications. In this protocol, we detail the stepwise procedures to conduct the LOCK (Long dsDNA with 3′-Overhangs mediated CRISPR Knock-in) method for gene-sized (~1–3 kb) KI in mammalian cells.
Graphical overview
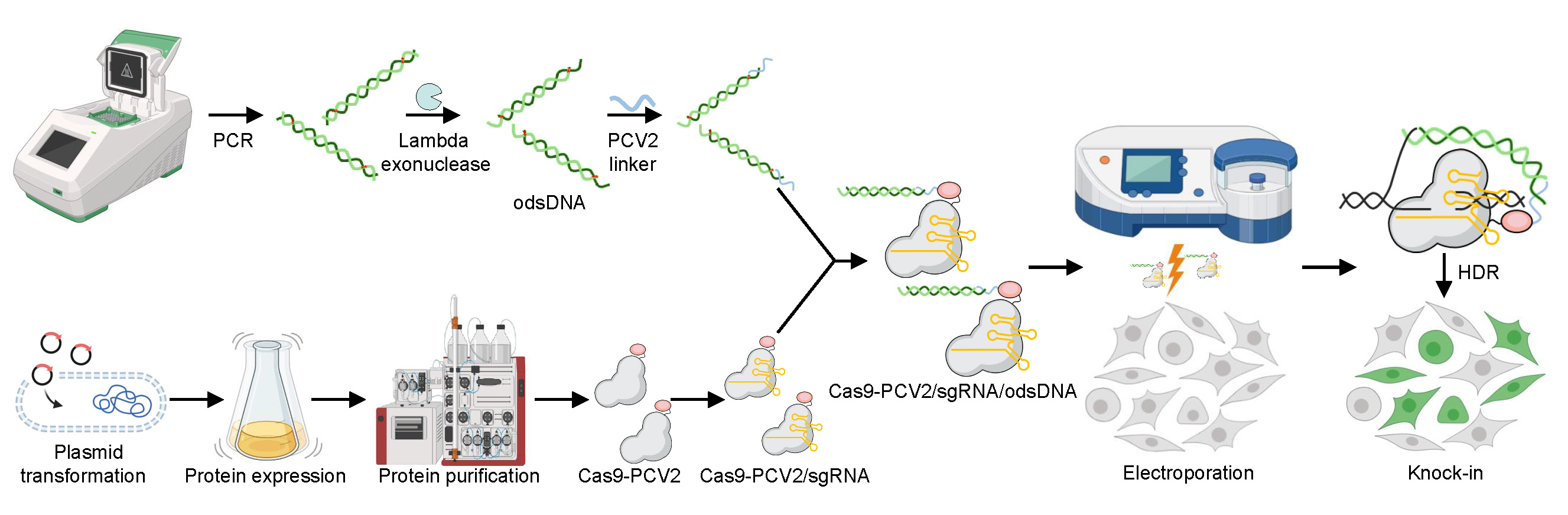
Improvement of large DNA fragment knock-in rates by attaching odsDNA donors to Cas9-PCV2 fusion protein
Background
The advent of CRISPR and its derived technologies have tremendously expanded the toolbox for genome manipulation for scientists in the fields of biomedical research and innovative biotechnological development. Compared with the ZFN or TALEN approaches, which are highly reliant on the specific recognition of the target genomic DNA (gDNA) sequence by specially engineered protein readers, the CRISPR-Cas9 method only necessitates a single crRNA (~20 nt) that uniquely recognizes and pairs with the specific gDNA locus (Hsu et al., 2014). This improvement makes genome-editing technique adoptable in any individual lab because of the low cost and simplified procedures. However, it is known that the precise integration of large donor DNA sequences into the host genome is dependent on homology-directed repair (HDR), which occurs at low frequencies, in mammalian cells. In addition, the double-strand break (DSB) sites induced by Cas9 cutting could happen at undesired genomic loci (off-target), and this might result in unexpected DSB repair outcomes, e.g., insertions/deletions (indels) and translocations, which are fundamentally detrimental to cellular health. It is, therefore, critical to increase the precise targeting efficiency while minimizing the off-target effects (Doudna, 2020).
In the past decade, the scientists have made great achievements in the improvement of the precise CRISPR-Cas9 knock-in (KI) efficiency. The phosphoric backbone of the donor DNA sequences is highly negatively charged, which naturally impedes their transportation across the cellular membrane, leading to the inaccessibility of exogenous DNA donors in the local DSB sites. This has been attributed as one of the reasons causing the low efficiencies of HDR-mediated KI. As such, versatile approaches have by far been devised to promote the nuclear delivery or the structural stability of the exogenous repair DNA donors (Yu et al., 2020). For instance, the nuclear entry of DNA donors could be promoted by adeno-associated viral (AAV) vectors, by Strep-biotin labeled tethering (Gu et al., 2018), or by chromatinized packaging (Cruz-Becerra and Kadonaga, 2020).
On the other hand, it is known that while the canonical nonhomologous end joining (c-NHEJ) repair is much more frequent at the CRISPR-Cas9-induced DSB sites, the repair pathway could be biased to the precise HDR repair outcome through artificially manipulating the cellular milieu (Maruyama et al., 2015). Indeed, accumulating examples have shown that the long ssDNA (lssDNA), as exogenous HDR donors, display superior advantages over the conventional double-strand DNA (dsDNA) donors, in terms of the HDR efficiency and off-target effect (Quadros et al., 2017; Shy et al., 2023). By leveraging the distinct advantages from both dsDNA and ssDNA donors, our previous study showed that a single hybrid “3′-overhang dsDNA” donor, termed odsDNA, has proved to significantly improve the HDR efficiencies by up to 5-fold increase with low off-target effect (Han et al., 2023). We named this Long dsDNA with 3′-Overhangs mediated CRISPR Knock-in, “LOCK” for short. In this protocol, we describe this easy technique with the stepwise procedures needed to be operational in any individual lab.
Materials and reagents
Materials
Pipette tips: 10, 200, and 1,000 μL (Yeasen, catalog numbers: 83010ES20, 83040ES20, and 83070ES08)
200 μL extended pipette tips, sterilized (Yeasen, catalog number: 83061ES50)
Pipettes: 2.5, 10, 20, 100, 200, and 1,000 μL (Eppendorf, catalog numbers: 3120000216, 3120000224, 3120000232, 3120000240, 3120000259, and 3120000267)
0.2 mL PCR strips of eight tubes (Yeasen, catalog number: 83602ES10)
1.5 mL microcentrifuge tube (Axygen, catalog number: MCT-150-C)
50 mL conical tube (Thermo Fisher, catalog number: 339652)
DNA oligonucleotide primers and PCV2 linker (Sangon Biotech)
6-well cell culture plate (BIOFIL, catalog number: TCP011006)
60 mm cell culture dish (BIOFIL, catalog number: TCD010060)
10 cm cell culture dish (BIOFIL, catalog number: TCD010100)
HEK293T cells (American Type Culture Collection, catalog number: CRL-3216)
DH5α competent cells (Sangon Biotech, catalog number: B528413)
Escherichia coli Rosetta 2 (DE3) strain (Millipore EMD, catalog number: 71397)
Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Invitrogen, catalog number: 15529019)
Tryptone (Gibco, catalog number: 211705)
Yeast extract (Gibco, catalog number: 211931)
Kanamycin monosulfate (Sangon Biotech, catalog number: A600286)
Agar (Thermo Fisher Scientific, catalog number: 22700025)
Terrific broth (Thermo Fisher Scientific, catalog number: 22711022)
Glycine (Invitrogen, catalog number: 15527013)
SDS (Sigma-Aldrich, catalog number: 436143)
NaCl (Sigma-Aldrich, catalog number: S9888)
KCl (Sigma-Aldrich, catalog number: P3911)
MgCl2 hexahydrate (Sigma-Aldrich, catalog number: M2670)
Tris (Invitrogen, catalog number: A32355)
HCl (Sigma-Aldrich, catalog number: 30721)
NaOH (Sigma-Aldrich, catalog number: 221465)
Imidazole (Sigma-Aldrich, catalog number: I2399)
UltraPure glycine (Invitrogen, catalog number: 15527013)
β-mercaptoethanol (Gibco, catalog number: 21985023)
EDTA (Thermo Fisher Scientific, catalog number: 17892)
HEPES (Sigma-Aldrich, catalog number: 54457)
TCEP (Thermo Fisher Scientific, catalog number: T2556)
Glycerol (Thermo Fisher Scientific, catalog number: 17904)
HisTrap fast flow, 5 mL (GE Healthcare, catalog number: GE17-5255-01)
Amicon Ultracel-100 regenerated cellulose membrane, 50 mL (Millipore, catalog number: UFC910008)
Amicon Ultracel-30 regenerated cellulose membrane, 4 mL (Millipore, catalog number: UFC803008)
0.22 μm syringe filter, PES membrane, 33 mm diameter (Millipore, catalog number: SLGPR33RS)
0.45 μm syringe filter, PES membrane, 33 mm diameter (Millipore, catalog number: SLHPR33RS)
Nalgene Rapid-Flow with PES 500 mL (Thermo Fisher Scientific, catalog number: 566-0020)
PD-10 desalting columns (GE Healthcare, catalog number: GE17-0851-01)
BeyoGel SDS-PAGE Precast Gel, Tris-Gly, 4%–20%, 12 wells (Beyotime, catalog number: P0057A)
Agarose (Thermo Fisher Scientific, catalog number: R0491)
GeneArt Precision gRNA Synthesis kit (Invitrogen, catalog number: A29377)
Invitrogen TrueCut Cas9 Protein v2 (Thermo Fisher Scientific, catalog number: A36497)
PARAFILM sealing film (Sigma-Aldrich, catalog number: HS234526B-1EA)
Kimwipes disposable wipers (Sigma-Aldrich, catalog number: Z188956)
Reagents
ddH2O, DEPC-treated water (Sangon Biotech, catalog number: B300592)
Lambda exonuclease (New England Biolabs, catalog number: M0262L)
GeneJET PCR Purification kit (Thermo Fisher Scientific, catalog number: K0702)
SpCas9 expression plasmid (Addgene, catalog number: 47327)
Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, catalog number: C11885500BT)
TrypLE express (Gibco, catalog number: 12604039)
DPBS (without Ca2+ or Mg2+) (Gibco, catalog number: 14190-144)
Fetal bovine serum (FBS) (Gibco, catalog number: 10091148)
Cell Line Nucleofector kit V (Lonza, catalog number: VCA-1003)
Q5 High-Fidelity 2× Master Mix (New England Biolabs, catalog number: M0492L)
50× TAE buffer (Sangon Biotech, catalog number: B548101)
6× Gel Loading Dye Purple (New England Biolabs, catalog number: B7024S)
SDS-PAGE Protein Loading Buffer, 5× (Beyotime, catalog number: P0286)
BeyoColor Prestained Color Protein Marker, 10–170 kDa (Beyotime, catalog number: P0077)
Imperial protein stain solution (Thermo Fisher Scientific, catalog number: 24615)
Annealing buffer for DNA Oligos, 5× (Beyotime, catalog number: D0251)
LB medium (see Recipes)
50 mg/mL kanamycin (see Recipes)
LB agar plate with kanamycin (see Recipes)
Terrific broth (TB) medium (see Recipes)
1 M IPTG (see Recipes)
Buffer A: start buffer (see Recipes)
Buffer B: elution buffer (see Recipes)
Buffer C: storage buffer (see Recipes)
Buffer D: cleavage buffer (see Recipes)
0.1 M NiSO4 (see Recipes)
0.1 M EDTA, 1 M NaCl (see Recipes)
1 M NaOH (see Recipes)
1 M HCl (see Recipes)
SDS-PAGE running buffer (see Recipes)
Low-EDTA TE buffer (see Recipes)
Recipes
LB medium (1,000 mL)
Reagent Final concentration Quantity Tryptone
Yeast extract
NaCl
10 g/L
5 g/L
10 g/L
10 g
5 g
10 g
ddH2O n/a ~980 mL Total n/a 1,000 mL Adjust the pH to 7.4 with NaOH. Autoclave at 121 °C for 15 min. Store at RT.
50 mg/mL kanamycin (10 mL)
Reagent Final concentration Quantity Kanamycin 50 mg/mL 0.5 g ddH2O n/a 10 mL Total n/a 10 mL Sterilize the solution with a 0.22 μm syringe filter. Store at 4 °C.
LB agar plate with kanamycin (1,000 mL)
Reagent Final concentration Quantity Tryptone
Yeast extract
NaCl
10 g/L
5 g/L
10 g/L
10 g
5 g
10 g
Agar 15 g/L 15 g 50 mg/mL Kanamycin 50 μg/mL 1 mL ddH2O n/a ~960 mL Total n/a 1,000 mL Adjust the pH to 7.4 with NaOH.
Autoclave at 121 °C for 15 min. Agar is added after sterilization.
Stir the medium on a stir plate and allow to cool to 30–40 °C.
Add 1 mL of 50 mg/mL kanamycin solution. Stir well.
Transfer 25 mL of the solution to 10 cm Petri dishes with a sterile pipette.
Allow the agar plates to cool and dry in a laminar flow hood for 30 min.
Store the plates at 4 °C in a plastic bag.
Terrific broth medium (1,000 mL)
Reagent Final concentration Quantity Terrific broth
Glycerol
47.6 g/L
0.4% (v/v)
47.6 g
4 mL
ddH2O n/a ~960 mL Total n/a 1,000 mL Adjust the pH to approximately 7.4 with NaOH. Autoclave at 121 °C for 15 min. Store at RT.
1 M IPTG (10 mL)
Reagent Final concentration Quantity IPTG 1 M 2.38 g ddH2O n/a ~9.8 mL Total n/a 10 mL Sterilize the solution with a 0.22 μm syringe filter. Store at -20 °C.
Buffer A: start buffer (1,000 mL)
Reagent Final concentration Quantity Tris
NaCl
20 mM
500 mM
2.4228 g
29.22 g
ddH2O n/a ~960 mL Total n/a 1,000 mL Adjust the pH to approximately 8.0 with HCl. Sterilize the solution with Nalgene Rapid-Flow. Store at 4 °C.
Buffer B: elution buffer (1,000 mL)
Reagent Final concentration Quantity Tris
NaCl
20 mM
500 mM
2.4228 g
29.22 g
imidazole 500 mM 34.04 g ddH2O n/a ~960 mL Total n/a 1,000 mL Adjust the pH to approximately 8.0 with HCl. Sterilize the solution with Nalgene Rapid-Flow. Store at 4 °C.
Buffer C: storage buffer (1,000 mL)
Reagent Final concentration Quantity Tris
KCl
20 mM
200 mM
2.4228 g
14.91 g
MgCl2·6H2O 10 mM 2.03 g Glycerol 10% (v/v) 100 mL ddH2O n/a ~860 mL Total n/a 1,000 mL Adjust the pH to approximately 8.0 with HCl. Sterilize the solution with Nalgene Rapid-Flow. Store at 4 °C.
Buffer D: cleavage buffer (1,000 mL)
Reagent Final concentration Quantity HEPES
KCl
20 mM
150 mM
4.77 g
11.18 g
MgCl2·6H2O 1 mM 0.2 g TCEP 1 mM 0.25 g Glycerol 10% (v/v) 100 mL ddH2O n/a ~860 mL Total n/a 1,000 mL Adjust the pH to approximately 8.0 with HCl. Sterilize the solution with Nalgene Rapid-Flow. Store at -20 °C.
0.1 M NiSO4 (100 mL)
Reagent Final concentration Quantity NiSO4·6H2O 0.1 M 2.63 g ddH2O n/a ~96 mL Total n/a 100 mL Sterilize the solution with a 0.22 μm syringe filter. Store at RT.
0.1 M EDTA, 1M NaCl (100 mL)
Reagent Final concentration Quantity EDTA 0.5 M 2.92 g NaCl 1 M 5.84 g ddH2O n/a ~96 mL Total n/a 100 mL Adjust the pH to 8.0 with NaOH. Sterilize the solution with a 0.22 μm syringe filter. Store at RT.
1 M NaOH (100 mL)
Reagent Final concentration Quantity NaOH 1 M 4 g ddH2O n/a ~96 mL Total n/a 100 mL Store at RT.
1 M HCl (120 mL)
Reagent Final concentration Quantity 12 M HCl 1 M 10 mL ddH2O n/a 110 mL Total n/a 120 mL Store at RT.
SDS-PAGE running buffer (1,000 mL)
Reagent Final concentration Quantity Tris
Glycine
125 mM
1.25 M
15.1 g
94 g
SDS 0.5% (w/v) 5 g ddH2O n/a ~800 mL Total n/a 1,000 mL Store at RT.
Low-EDTA TE buffer, 1× (for DNA) (100 mL)
Reagent Final concentration Quantity Tris
EDTA
10 mM
0.2 mM
0.1211 g
0.0012 g
ddH2O n/a ~100 mL Total n/a 100 mL Adjust the pH to 8.0 with HCl. Sterilize the solution with a 0.22 μm syringe filter. Store at RT.
Equipment
Amaxa Nucleofector IIb device (Lonza, model: AAB-1001)
BD Accuri C6 flow cytometer (BD Biosciences, catalog number: 23432)
Heracell VIOS 160i CO2 copper chamber incubator (Thermo Fisher Scientific, catalog number: 51030476)
Biological safety cabinet (Esco Airstream, model: AB2-4S8-CN)
Sonicator Q125 (Qsonica, model: Q125-220)
1/4 diameter sonica probe (Qsonica, catalog number: 4435)
Protein purification system (AKTA pure, catalog number: 29046665)
Fluorescence spectrophotometer F-7000 (HITACHI, model: F-7000)
IX71 inverted microscope (Olympus, model: IX71)
Snowflake ice machine (XUEKE, model: IMS-20)
Water bath (EYELA, model: NTT-2200)
Mini gel DNA electrophoresis system (Thermo Fisher, model: B1)
Gel imaging system (Azure Biosystems, model: C300)
Veriti 96- Well PCR thermal cycler (Applied Biosystems, catalog number: 4375786)
TAdvanced Twin PCR thermocycler (Biometra, catalog number: 2070212)
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, model: ND-2000)
pH meter (Mettle Toledo, catalog number: 51302807)
Milli-Q water purification system (Millipore, model: MP0024)
Eppendorf centrifuge 5424 R (Eppendorf, catalog number: 5404000090)
Eppendorf high-speed refrigerated centrifuge 5910 R (Eppendorf, catalog number: 5942000598)
Thermo shaker (Yeasen, catalog number: 80440ES03)
Software
Prism v8, for statistics and data visualization (GraphPad, version 8.0.1.244)
Snapgene, view DNA sequence and primer design (version 4.2.4)
NUPACK: Nucleic Acid Package (https://nupack.org/)
Tm Calculator (version 1.16.5, https://tmcalculator.neb.com/#!/main)
CHOPCHOP, a web-based tool for sgRNA design. The website is for non-profit and academic use only (https://chopchop.cbu.uib.no/)
Expasy, conversion of DNA sequence to amino acid sequence (https://web.expasy.org/translate/)
AAT Bioquest: Protein Concentration Calculator (https://www.aatbio.com/tools/calculate-protein-concentration)
NEBioCalculator (https://nebiocalculator.neb.com/#!/dsdnaamt)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Han, W., Liang, H. and Bao, J. (2023). Efficient Large DNA Fragment Knock-in by Long dsDNA with 3′-Overhangs Mediated CRISPR Knock-in (LOCK) in Mammalian Cells. Bio-protocol 13(20): e4853. DOI: 10.21769/BioProtoc.4853.
分类
分子生物学 > DNA > DNA 修饰
细胞生物学 > 细胞工程 > CRISPR-cas9
癌症生物学 > 通用技术 > 分子生物学技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











