- EN - English
- CN - 中文
A Protocol to Retrieve and Curate Spatial and Climatic Data from Online Biodiversity Databases Using R
使用 R 从在线生物多样性数据库检索和管理空间和气候数据的实验方案
发布: 2023年10月20日第13卷第20期 DOI: 10.21769/BioProtoc.4847 浏览次数: 1573
评审: Prashanth N SuravajhalaJeff W Bizzaro
Abstract
Ecological and evolutionary studies often require high quality biodiversity data. This information is readily available through the many online databases that have compiled biodiversity data from herbaria, museums, and human observations. However, the process of preparing this information for analysis is complex and time consuming. In this study, we have developed a protocol in R language to process spatial data (download, merge, clean, and correct) and extract climatic data, using some genera of the ginseng family (Araliaceae) as an example. The protocol provides an automated way to process spatial and climatic data for numerous taxa independently and from multiple online databases. The script uses GBIF, BIEN, and WorldClim as the online data sources, but can be easily adapted to include other online databases. The script also uses genera as the sampling unit but provides a way to use species as the target. The cleaning process includes a filter to remove occurrences outside the natural range of the taxa, gardens, and other human environments, as well as erroneous locations and aspatial correction for misplaced occurrences (i.e., occurrences within a distance buffer from the coastal boundary). Additionally, each step of the protocol can be run independently. Thus, the protocol can begin with data cleaning, if the database has already been compiled, or with climatic data extraction, if the database has already been parsed. Each line of the R script is commented so that it can also be run by users with little knowledge of R.
Keywords: Ecology (生态学)Background
Our knowledge of species distributions is central to biogeographers but also to phylogeneticists and ecologists. Indeed, species distributions are needed to perform phylogenetic climate reconstructions, species niche characterizations, or species distribution models, and to address many evolutionary questions. However, obtaining accurate spatial information on species distributions requires occurrence databases of good quality, with high geographical coverage, which are difficult to obtain.
The main sources of geographical information are field inventories and biodiversity collections (museums and herbaria), for which accessibility has been a serious limitation until recently. The digitization efforts of recent decades have facilitated access to vast amounts of biodiversity data previously scattered in different institutions around the world, through online databases such as the Global Biodiversity Information Facility (GBIF; GBIF.org, 2021). As a result, we now have an unprecedented opportunity to benefit from centuries worth of naturalist observations from all over the world. However, the use of this valuable information is limited by persistent knowledge gaps and technical limitations. On the one hand, our knowledge of species distributions is still poor, biased, or imprecise (Hortal et al., 2007), and this is reflected in the information collected in biodiversity databases, which is not consistent across lineages or across regions. These biases result in some groups of organisms and regions of the world having scarce information while others have large amounts of data (Hortal and Lobo, 2005). On the other hand, the complexity of the process of parsing and preparing online data for analysis is high. For example, it is common for online repositories to contain records with imprecise or erroneous spatial information (such as terrestrial organisms with records in the sea) or with outdated taxonomic nomenclature (Soberón and Peterson, 2004). Therefore, every study based on online data requires a first step of cleaning and parsing to remove or minimize the impact of these sources of uncertainty (persistent knowledge gaps and technical limitations) on further analysis (Hortal et al., 2007).
In parallel with the international digitization efforts of the last decades, several methods and pipelines have been designed to deal with these sources of uncertainty and to simplify the different steps of working with online biodiversity data. Some of the most relevant protocols have been developed in R (R Core Team, 2018) and include geographic, taxonomic, or temporal data cleaning (see for example: Biogeo, Robertson et al., 2016; SpeciesGeoCorder, Töpel et al., 2016; CoordinateCleaner, Zizka et al., 2019; bRacatus, Arlé et al., 2021; BDcleaner, Jin and Yang, 2020; plantR, Lima et al., 2021). However, none of them address the uncertainty introduced by both the spatial knowledge gaps and the technical limitations. Moreover, most of them focus on one or a few steps of the process. Thus, to complete the process (from the initial download of occurrences to the climatic data extraction of the cleaned and parsed spatial database) users have to deal with different protocols, some of which require programming skills or a deep R background.
The R protocol that we present here is designed to produce reliable databases of species occurrences and climate data from online repositories. It provides an automatic procedure for dealing with the most common sources of spatial uncertainty in online biodiversity databases. It also includes an automatic script to run each sample (species, genus, family, etc.) separately, allowing for an easy and fast way to process hierarchical databases. The script also includes a post-processing code to run after the spatial pipeline and extract the climatic data. The protocol describes a step-by-step guide on how to download, parse, clean, and merge spatial and climatic data from three online databases (Figure 1; WorldClim, Fick and Hijmans, 2017; BIEN, Maitner, 2020; GBIF, GBIF.org, 2021). Moreover, the protocol can be easily adapted to include any other online biodiversity database that may be of interest. The cleaning steps include how to automatically update nomenclatural information, identify, and remove records outside the natural distribution of taxa, records from gardens and other human environments, or geographically inaccurate records. To explain the protocol, we used the AsianPalmate Group (AsPG) of Araliaceae as a case study, using genera as a sample unit. To speed up the implementation, we selected 16 of the AsPG genera. The selection of genera was made to exhibit uneven spatial information across genera and across areas of the world. This approach aimed to address the issue derived from knowledge gaps as a source of spatial uncertainty. Additionally, the chosen genera are largely affected by erroneous and misplaced records, serving to tackle the issue arising from technical limitations as a source of spatial uncertainty.
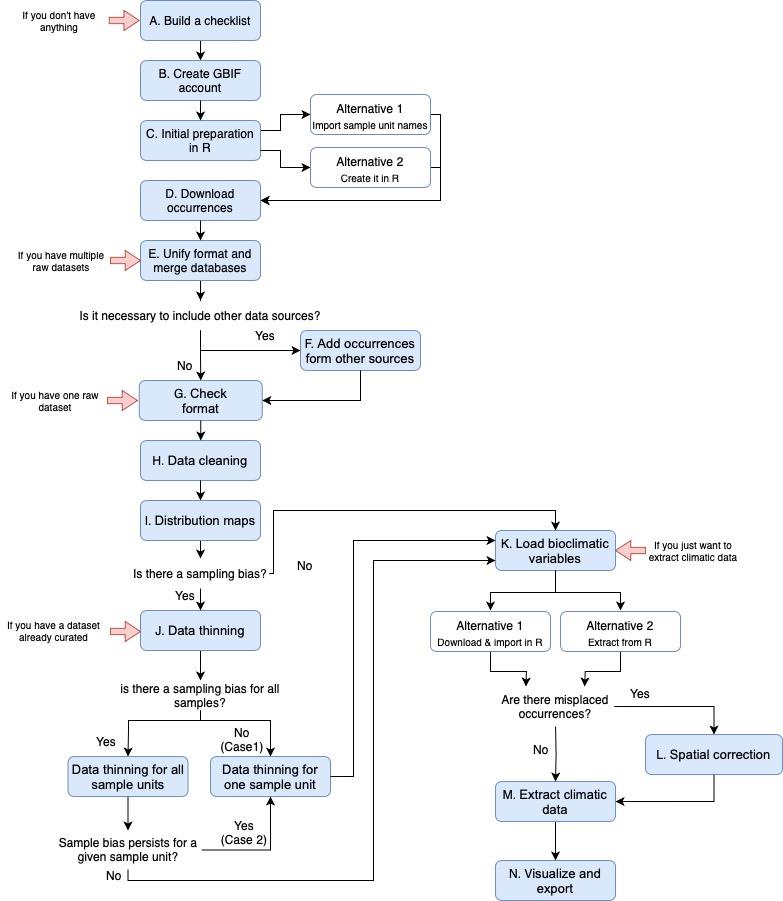
Figure 1. Workflow for the entire protocol pipeline. It includes all the steps and alternatives described in the protocol. Each red arrow represents the steps of the protocol that we can start with, depending on the data available.
In summary, the main advantages of this protocol are that it: (1) can be applied to all groups of organisms (as long as they have information available in GBIF or BIEN databases) and at any taxonomic rank, not only at the species level; (2) provides an automatic way to handle hierarchical databases, which is very helpful when studying highly diversified groups (genera with a high number of species, families with a high number of genera, etc.); (3) provides a complete pipeline from spatial data download (including merging multiple databases) to climatic data extraction; (4) deals with uncertainties arising from technical limitations (such as incorrect records), but also with the uncertainties arising from persistent knowledge gaps (such as spatial biases in different parts of the world and across lineages); (5) provides an easy way to filter out records outside the natural range of the taxa; (6) applies a spatial correction for erroneous occurrences outside the coastal boundary; (7) includes independent steps for each part of the process that can be run separately; and (8) can be easily used and modified by any types of users regardless of their skills, knowledge, or background on R, because it is accompanied by instructions to guide the user.
Equipment
Computer with Microsoft® Windows® 10 education, KUBUNTU 22.04, or Mac® OS X® 12.6 operating systems and versions.
Software
R version 4.3.1 (https://r-project.org/)
Packages: "BIEN", "countrycode", "data.table", "devtools", "dpyr", "plyr", "raster", "readr", "rgbif", "rgdal", "spocc", "spThin", "SEEG-Oxford/seegSDM" and "tidyr".
RStudio version 2023.06.0+421 (https://rstudio.com/products/rstudio/)
The use of RStudio is optional. RStudio is an interface that improves the use of R.
Databases
POWO (https://powo.science.kew.org)
GBIF (https://www.gbif.org/)
WorldClim (https://www.worldclim.org/)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Coca-de-la-Iglesia, M., Valcárcel, V. and Medina, N. G. (2023). A Protocol to Retrieve and Curate Spatial and Climatic Data from Online Biodiversity Databases Using R. Bio-protocol 13(20): e4847. DOI: 10.21769/BioProtoc.4847.
分类
生物信息学与计算生物学
环境生物学 > 生态系统
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
提问指南
+ 问题描述
写下详细的问题描述,包括所有有助于他人回答您问题的信息(例如实验过程、条件和相关图像等)。
Share
Bluesky
X
Copy link









