- EN - English
- CN - 中文
Doxycycline-inducible Expression of Proteins at Near-endogenous Levels in Mammalian Cells Using the Sleeping Beauty Transposon System
使用睡美人转座子系统在哺乳动物细胞中以近内源水平表达多西环素诱导的蛋白质
发布: 2023年10月20日第13卷第20期 DOI: 10.21769/BioProtoc.4846 浏览次数: 2225
评审: Gal HaimovichKeisuke TabataAnonymous reviewer(s)
Abstract
The function of a protein within a cell critically depends on its interaction with other proteins as well as its subcellular localization. The expression of mutants of a particular protein that have selective perturbation of specific protein interaction motifs is a very useful strategy for resolving a protein’s mechanism of action in a cellular process. In addition, expression of fluorescent protein fusions is a key strategy for determining the subcellular localization of a protein. These strategies require tight regulation to avoid potential alterations in protein interactions or localizations that can result from protein overexpression. Previous work led to the development of a Sleeping Beauty transposon system that allows doxycycline-inducible expression of protein mutants or fusions; titration of doxycycline allows expression of protein fusions or mutants at near endogenous levels. When used in combination with siRNA gene silencing, this strategy allows for knockdown-rescue experiments to assess the function of specific protein mutants. In this protocol, we describe the use of this Sleeping Beauty strategy for expression of eGFP fusion or mutant proteins in ARPE-19 and MDA-MB-231 cells. This includes design of expression plasmids, transfection, and selection to obtain stable engineered cells, as well as doxycycline treatment for controlled induction of protein expression, either alone or in combination with siRNA silencing for knockdown-rescue experiments. This strategy is advantageous as it allows rapid generation of stable cells for controlled protein expression, suitable for functional studies that require knockdown-rescue as well as various forms of live cell fluorescence imaging.
Key features
• Highly versatile doxycycline-inducible expression system that can be used in various mammalian cell lines.
• Stable integration of transgene allows for sustained and stable expression.
• Titration of doxycycline levels allows expression of transgene at near endogenous levels.
Background
Cell biology studies often require the expression of exogenous or modified proteins (through the use of transgenes) in cells in culture or other model systems, including expression of specific mutants that each selectively perturb a single aspect of that protein’s function. In addition, the study of protein localization often requires expression of the protein of interest fused to a fluorescent protein such as eGFP (Snapp, 2005) or to an enzyme suitable for proximity biotinylation (Gingras et al., 2019). Hence, optimal strategies for expression of transgenes are essential for a wide range of cell biology approaches.
Transient transfection to introduce plasmids encoding these various transgenes into mammalian cells in culture are commonly used for this purpose. However, this strategy can lead to artificial localization or function of the expressed proteins, as the level of expression of the exogenous proteins using this approach often dramatically exceeds that of the endogenous. Furthermore, transient transfection often leads to expression restricted to a minor fraction of cells (5%–20%), which limits the use of this method for cell population–based assays. In contrast, retroviral and lentiviral systems allow for efficient integration of a cassette for expression of a transgene into the genome. When combined with a selection stage, this approach allows for expression of the transgene in a large majority of cells in a population, as well as relatively stable expression levels of the transgene over time. However, the utility of lentiviral or retroviral strategies may be limited by size constraints for the transgene imposed by the packaging process, the time required to generate viral particles, as well as the need for specific biosafety protocols for work with these virus-like particles.
The modification of endogenous genes with CRISPR/Cas9 has also emerged as a compelling strategy to allow expression of specific mutants or fusions of a specific gene from its endogenous promoter, thus avoiding artifacts of protein overexpression (Bukhari and Müller, 2019). While powerful, this strategy also has potential limitations. Generation of loss-of-function mutants of a particular gene using CRISPR/Cas9 involves passaging of these modified cells and may lead to cellular adaptation that may limit the study of the direct effects of the protein being studied. Furthermore, many genes are present in multiple copies in many cultured cell lines, in particular aneuploid cell lines such as those derived from tumors (e.g., the breast cancer cell line MDA-MB-231 used here). Hence, it can be challenging to use CRISPR/Cas9 to modify all copies of a gene within the genome of these cells in culture, limiting the use of this approach for expression of loss-of-function mutants.
The use of transposons for stable integration of transgenes into the genome of cultured cells is an effective strategy that parallels the use of viral transduction strategies, without the need for packaging of transgene material into viral particles. The Sleeping Beauty transposon method enables stable integration of a transgene found within a vector containing inverted terminal repeats (ITRs) when co-transfected into cells with a vector encoding a transposase enzyme. This approach can be used in combination with selection to achieve integration of the transgene into a large majority of cells in a population (Kowarz et al., 2015).
While the Sleeping Beauty system has been used extensively, a re-engineered transposase enzyme with enhanced activity—SB100X (Mátés et al., 2009)—is ideally suited for this approach, and achieves highly efficient and random integration into TA dinucleotide sites within the target cell genome (30,000 such sites in the human genome) (Kowarz et al., 2015). Kowarz & col. developed a suite of plasmids for use of the Sleeping Beauty transposon system that includes plasmids to allow either inducible (pSBtet) or constitutive (e.g., pSBbi) expression and a range of fluorescent protein and antibiotic selection markers and demonstrated the use of these strategies for stable gene expression in HeLa and HEK293T cells (Kowarz et al., 2015). The pSBtet series of plasmids developed by Kowarz & col. allows doxycycline-inducible expression of a transgene alongside constitutive expression of one of several selection markers (puromycin, hygromycin, neomycin, or blasticidin) and a fluorescent protein (BFP, eGFP, or RFP). We focus here on the use of pSBtet-BP (constitutive expression of BFP and puromycin selection, Figure 1A). Notably, the selection marker, fluorescent protein, and reverse tetracycline transactivator (rtTA) in the pSBtet plasmids are expressed as a self-cleaving polyprotein (Figure 1A).
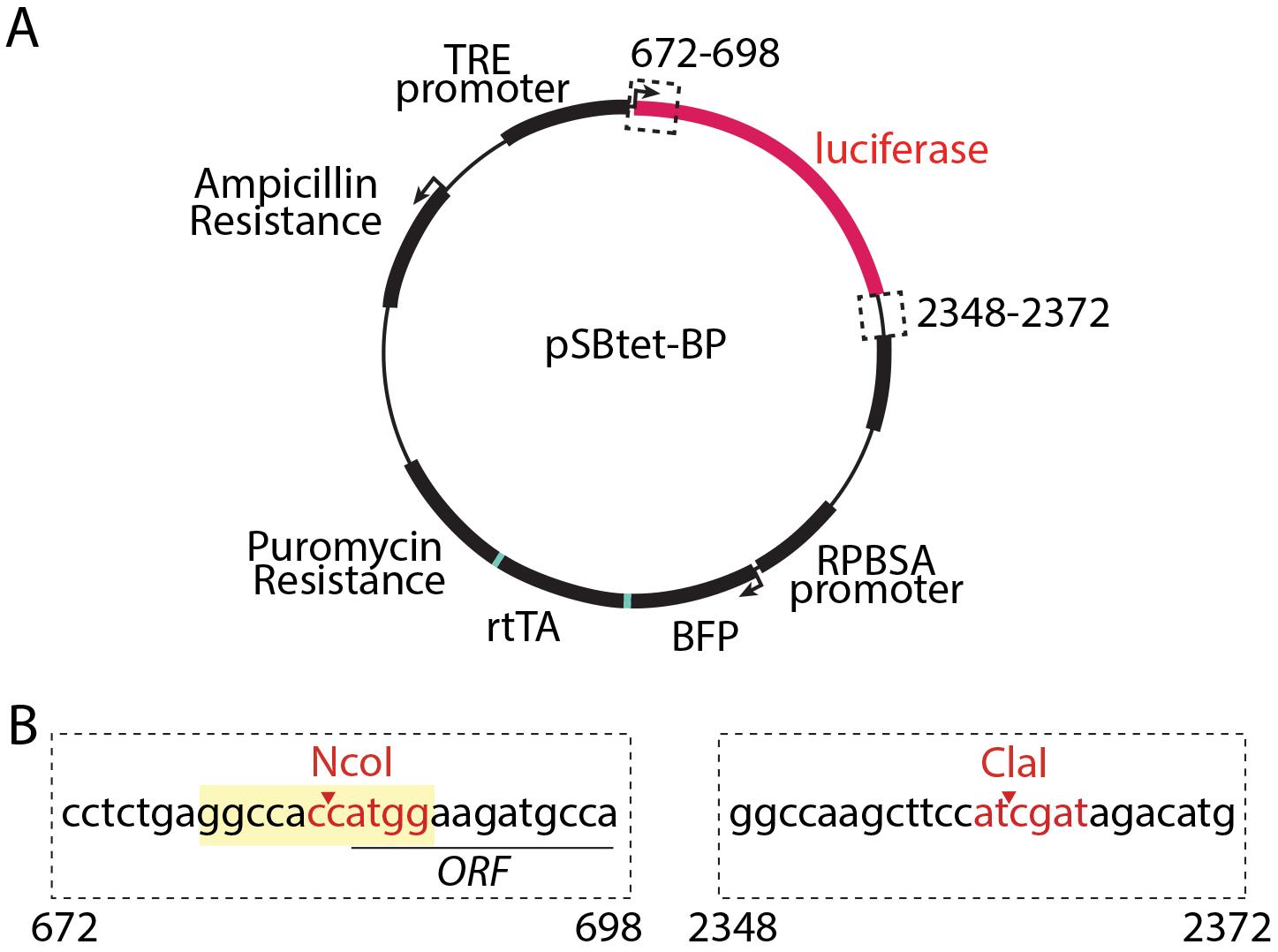
Figure 1. pSBtet-BP plasmid and cloning strategy. (A) Map of the pSBtet-BP plasmid as obtained from Addgene, plasmid number 60496 and developed by Kowarz & col. (Kowarz et al., 2015). Shown are the Tetracycline responsive element (TRE) promoter, the open reading frame encoding luciferase (which is replaced by the transgene of interest during the cloning stage, Procedure A), as well as the expression of a polyprotein from the constitutive synthetic RPBSA promoter; this polyprotein undergoes cleavage to generate blue fluorescent protein (BFP), reverse tetracycline transactivator (rtTA), and puromycin resistance protein. Also shown is the expression of ampicillin resistance for bacterial selection. Also shown (dashed line boxes) are the regions relevant to subcloning of the transgene, as described in Procedure A. (B) Shown is the cloning strategy for excision of the luciferase-encoding sequence and subcloning of the transgene of interest using NcoI and ClaI sites within the pSBtet-BP plasmid. NcoI and ClaI cut sites are shown in red text; the Kozak sequence is shown in yellow and the open reading frame (ORF) of the luciferase is underlined.
We recently used the Sleeping Beauty transposon strategy developed by Kowarz & col. (Kowarz et al., 2015) in ARPE-19, MDA-MB-231, and SUM149-PT cells for generation of stable cells that allow doxycycline-inducible expression of protein fusions or mutants (Cabral-Dias et al., 2022; Rahmani et al., 2023; Sugiyama et al., 2023) or short hairpin RNA (shRNA) (Lo et al., 2023). Here, we first describe subcloning of a transgene of interest into the pSBtet plasmid (Procedure A). We also note that the use of these Sleeping Beauty plasmids requires an initial transfection and selection process that is unique to some cell lines. We thus report the protocol optimized for the generation of stable cells in ARPE-19 and MDA-MB-231 (Procedure B). Finally, the use of doxycycline for expression of a transgene by the Sleeping Beauty strategy can allow for control of expression to approximate the endogenous level of expression of the protein. This is achieved through titration of doxycycline, allowing determination of an optimal range of doxycycline for each stable cell line generated. This approach allows expression of the transgene at near-endogenous levels, either alone or in combination with siRNA gene silencing of the endogenous protein for knockdown-rescue approaches (Cabral-Dias et al., 2022; Rahmani et al., 2023; Sugiyama et al., 2023). Thus, we also describe the use of doxycycline to induce expression of the protein of interest, either alone to determine optimal doxycycline condition for expression of the transgene (Procedure C), or in combination with siRNA gene silencing of the endogenous protein for knockdown-rescue approaches (Procedure D).
Materials and reagents
Restriction enzymes, e.g., NcoI and ClaI (New England Biolabs, catalog numbers: R0193L and R0197L, respectively)
Competent DH5α E. coli (Thermo Fisher, catalog number: 18265017)
D-MEM/F-12 (1×), liquid 1:1, with L-glutamine and HEPES buffer, 500 mL (Gibco, catalog number: 11330032)
RPMI-1640 medium, liquid; with L-glutamine and sodium bicarbonate, 500 mL (Sigma-Aldrich, catalog number: R8758-500ML)
Tissue culture flasks, 75 cm sq (250 mL, vented, polystyrene) (Sarstedt, catalog number: 83.3911.002)
6-well tissue culture plates, sterile, polystyrene (Sarstedt, catalog number: 86.1254.001)
Fetal bovine serum (Canada), 500 mL (Thermo Fisher Scientific, catalog number: 12483020)
Tetracycline-Free FBS (Wisent, catalog number: 081-150)
Penicillin-Streptomycin liquid 100 mL (Gibco, catalog number: 15070063)
Trypsin-EDTA (0.25% Trypsin with EDTA 1×, 500 mL (Gibco, catalog number: 25200072)
Dulbecco’s phosphate-buffered saline (dPBS), without calcium chloride and magnesium chloride, liquid, 500 mL (Sigma-Aldrich, catalog number: D8537-500ML)
Multiple PCR microtube, 0.5 mL (thin wall, neutral) (Sarstedt, catalog number: 72.735.002)
Ampicillin, sodium salt, 25 g (BioShop, catalog number: AMP201.25)
Chloramphenicol, 25 g (BioShop, catalog number: CLR201.25)
LB Broth (Miller), liquid microbial growth medium (Sigma-Aldrich, catalog number: L2542-500ML)
NucleoBond Xtra Midi EF, Midi kit for endotoxin-free plasmid DNA (Macherey-Nagel, catalog number: 740420.10)
FuGENE HD Transfection Reagent, 1 mL (Promega, catalog number: E2311)
Opti-MEM reduced serum medium (Thermo Fisher Scientific, catalog number: 31985070)
Puromycin dihydrochloride (Sigma-Aldrich, catalog number: P7255-25MG)
pCMV(CAT)T7-SB100 plasmid (Addgene, Plasmid #34879)
pSBtet-BP plasmid (Addgene, Plasmid #60496)
Note: Various other plasmids described in Kowarz et al. (2015) allow alternatives for generation of stable cells to express the transgene using the Sleeping Beauty transposon system.
siRNA in RNAse-free water reconstituted at 20 μM
Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific, catalog number: 13778150)
MDA-MB-231 cells (ATCC, catalog number: HTB-26)
ARPE-19 cells (ATCC, catalog number: CRL-2302)
ARPE-19 growth medium (see Recipes)
Tet-free ARPE-19 growth medium (see Recipes)
MDA-MB-231 growth medium (see Recipes)
Tet-free MDA-MB-231 growth medium (see Recipes)
Equipment
Shaking 37 °C platform incubator (Thermo Fisher Scientific, model: SHKE6000)
Cell incubator set to 37 °C and 5% CO2 (Thermo Fisher Scientific, model: HERAcell 150i)
Centrifuge (unrefrigerated) for 50 mL Falcon tubes (Thermo Fisher Scientific, model: Sorval ST16R)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zak, K. and Antonescu, C. N. (2023). Doxycycline-inducible Expression of Proteins at Near-endogenous Levels in Mammalian Cells Using the Sleeping Beauty Transposon System. Bio-protocol 13(20): e4846. DOI: 10.21769/BioProtoc.4846.
- Cabral-Dias, R., Lucarelli, S., Zak, K., Rahmani, S., Judge, G., Abousawan, J., DiGiovanni, L. F., Vural, D., Anderson, K. E., Sugiyama, M. G., et al. (2022). Fyn and TOM1L1 are recruited to clathrin-coated pits and regulate Akt signaling. J. Cell Biol. 221(4): e201808181.
分类
分子生物学 > 蛋白质 > 蛋白质-蛋白质相互作用
分子生物学 > 蛋白质 > 表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
提问指南
+ 问题描述
写下详细的问题描述,包括所有有助于他人回答您问题的信息(例如实验过程、条件和相关图像等)。
Share
Bluesky
X
Copy link