- EN - English
- CN - 中文
Testing for Allele-specific Expression from Human Brain Samples
测试人脑样本的等位基因特异性表达
发布: 2023年10月05日第13卷第19期 DOI: 10.21769/BioProtoc.4832 浏览次数: 1900
评审: Geoffrey C. Y. LauZhengrong YuanUte Angelika HoffmannAnonymous reviewer(s)

相关实验方案
基于 rAAV-α-Syn 与 α-Syn 预成纤维共同构建的帕金森病一体化小鼠模型
Santhosh Kumar Subramanya [...] Poonam Thakur
2025年12月05日 1425 阅读
Abstract
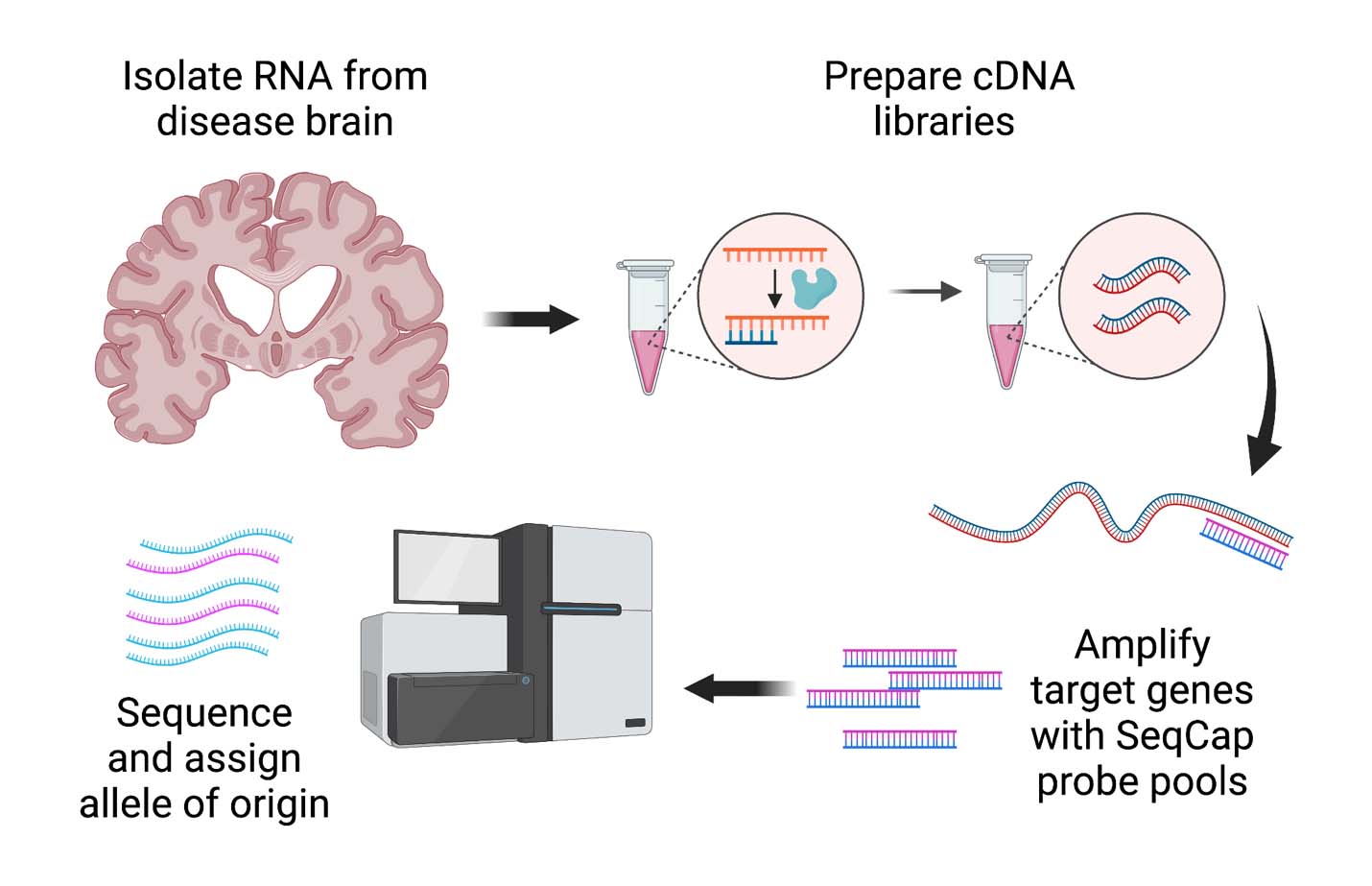
Many single nucleotide polymorphisms (SNPs) identified by genome-wide association studies exert their effects on disease risk as expression quantitative trait loci (eQTL) via allele-specific expression (ASE). While databases for probing eQTLs in tissues from normal individuals exist, one may wish to ascertain eQTLs or ASE in specific tissues or disease-states not characterized in these databases. Here, we present a protocol to assess ASE of two possible target genes (GPNMB and KLHL7) of a known genome-wide association study (GWAS) Parkinson’s disease (PD) risk locus in postmortem human brain tissue from PD and neurologically normal individuals. This was done using a sequence of RNA isolation, cDNA library generation, enrichment for transcripts of interest using customizable cDNA capture probes, paired-end RNA sequencing, and subsequent analysis. This method provides increased sensitivity relative to traditional bulk RNAseq-based and a blueprint that can be extended to the study of other genes, tissues, and disease states.
Key features
• Analysis of GPNMB allele-specific expression (ASE) in brain lysates from cognitively normal controls (NC) and Parkinson’s disease (PD) individuals.
• Builds on the ASE protocol of Mayba et al. (2014) and extends application from cells to human tissue.
• Increased sensitivity by enrichment for desired transcript via RNA CaptureSeq (Mercer et al., 2014).
• Optimized for human brain lysates from cingulate gyrus, caudate nucleus, and cerebellum.
Graphical overview
Background
Most single nucleotide polymorphisms (SNPs) associated with disease traits by genome-wide association studies (GWAS) are in non-coding regions of the genome. One way in which these SNPs may influence disease risk is via effects on expression of nearby genes as expression quantitative trait loci (eQTL). Public resources such as the Genotype-Tissue Expression (GTEx) Project allow scientists to easily query large datasets from multiple tissues from humans without disease for eQTL relationships between SNP variants and specific RNA transcripts in various organ tissues from healthy individuals. However, in many cases, investigators may want to understand whether an eQTL relationship extends to specific tissues from specific individuals; for example, either in tissues not queried by these databases or in individuals with a disease condition. Because the mechanistic basis for many eQTL effects is allele-specific expression (ASE), we developed a protocol for assaying ASE in brain tissue from human subjects with Parkinson’s disease (PD) who are heterozygous for the eQTL SNP in question, affecting the target gene GPNMB. This protocol builds on the ASE method described by Mayba et al. (2014) by enriching for the desired transcripts via an RNA CaptureSeq (Mercer et al., 2014) step and by adapting the protocol to assay human brain tissue as opposed to cultured cells. Furthermore, our protocol can be adapted to multiple complex traits and tissues to advance our mechanistic understanding of many non-coding risk variants with eQTL effects.
Materials and reagents
Biological materials
Human brain samples (acquired from University of Pennsylvania’s CNDR Brain Bank)
Reagents
TriZol (Invitrogen, catalog number: 15596026)
Qiagen RNeasy kit (Qiagen, catalog number: 74104)
Agilent Bioanalyzer RNA 6000 Nano Kit (Agilent, catalog number: 5067-1511)
Agilent Bioanalyzer DNA 1000 Nano (Agilent, catalog number: 5067-1504)
Agencourt AMPure XP beads (Agencourt, catalog number: A63881)
KAPA RNA HyperPrep Kit (KAPA/Roche Sequencing solutions, catalog number: KK8540)
Roche SeqCap RNA probes (Roche, custom designed, see Procedure below)
Roche SeqCap EZ Accessory kit v2 (Roche, catalog number: 07145594001)
Roche SeqCap EZ Hybridization and Wash Kit (Roche, catalog number: 05634261001)
Ethanol (Decon Labs, Inc., catalog number: 2716)
Water for RNA applications (Fisher bioreagents, catalog number: BP5611)
Laboratory supplies
1.5 mL plastic tubes (Eppendorf, catalog number: 022431021)
0.2 mL PCR tubes (Denville, catalog number: C18098-4)
Equipment
Bioanalyzer (Agilent 2100 Bioanalyzer, catalog number: G2939BA)
Thermocycler (Applied Biosystems, Veriti 96-well Thermal Cycler, catalog number: 4375786)
Sequencer (Illumina HiSeq 2500, catalog number: SY-401-2501)
Dyna-mag2 magnet (Thermo Fisher, catalog number: 12321D)
Vortex mixer (Fisherbrand, Brand, catalog number: 02-215-365)
Microcentrifuge (Eppendorf Model 5424, catalog number: 022620401)
NanoDrop or similar spectrophotometer
Software and datasets
RStudio
FastQC
Trimmomatic (Version 032)
STAR
WASP
VariantAnnotation
TxDb.Hsapiens.UCSC.hg19.knownGene
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Diaz-Ortiz, M. E., Jain, N., Gallagher, M. D., Posavi, M., Unger, T. L. and Chen-Plotkin, A. S. (2023). Testing for Allele-specific Expression from Human Brain Samples. Bio-protocol 13(19): e4832. DOI: 10.21769/BioProtoc.4832.
- Diaz-Ortiz, M. E., Seo, Y., Posavi, M., Carceles Cordon, M., Clark, E., Jain, N., Charan, R., Gallagher, M. D., Unger, T. L., Amari, N., et al. (2022). GPNMB confers risk for Parkinson’s disease through interaction with α-synuclein. Science 377(6608): eabk0637.
分类
神经科学 > 神经系统疾病 > 帕金森氏症
分子生物学 > RNA > RNA 测序
系统生物学 > 基因组学 > 测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link