- EN - English
- CN - 中文
Controlled Level of Contamination Coupled to Deep Sequencing (CoLoC-seq) Probes the Global Localisation Topology of Organelle Transcriptomes
控制污染水平与深度测序 (CoLoC-seq) 相结合以探索细胞器转录组的全局定位拓扑
发布: 2023年09月20日第13卷第18期 DOI: 10.21769/BioProtoc.4820 浏览次数: 2693
评审: Alessandro DidonnaMarina Sánchez PetidierRajesh D Gunage
Abstract
Information on RNA localisation is essential for understanding physiological and pathological processes, such as gene expression, cell reprogramming, host–pathogen interactions, and signalling pathways involving RNA transactions at the level of membrane-less or membrane-bounded organelles and extracellular vesicles. In many cases, it is important to assess the topology of RNA localisation, i.e., to distinguish the transcripts encapsulated within an organelle of interest from those merely attached to its surface. This allows establishing which RNAs can, in principle, engage in local molecular interactions and which are prevented from interacting by membranes or other physical barriers. The most widely used techniques interrogating RNA localisation topology are based on the treatment of isolated organelles with RNases with subsequent identification of the surviving transcripts by northern blotting, qRT-PCR, or RNA-seq. However, this approach produces incoherent results and many false positives. Here, we describe Controlled Level of Contamination coupled to deep sequencing (CoLoC-seq), a more refined subcellular transcriptomics approach that overcomes these pitfalls. CoLoC-seq starts by the purification of organelles of interest. They are then either left intact or lysed and subjected to a gradient of RNase concentrations to produce unique RNA degradation dynamics profiles, which can be monitored by northern blotting or RNA-seq. Through straightforward mathematical modelling, CoLoC-seq distinguishes true membrane-enveloped transcripts from degradable and non-degradable contaminants of any abundance. The method has been implemented in the mitochondria of HEK293 cells, where it outperformed alternative subcellular transcriptomics approaches. It is applicable to other membrane-bounded organelles, e.g., plastids, single-membrane organelles of the vesicular system, extracellular vesicles, or viral particles.
Key features
• Tested on human mitochondria; potentially applicable to cell cultures, non-model organisms, extracellular vesicles, enveloped viruses, tissues; does not require genetic manipulations or highly pure organelles.
• In the case of human cells, the required amount of starting material is ~2,500 cm2 of 80% confluent cells (or ~3 × 108 HEK293 cells).
• CoLoC-seq implements a special RNA-seq strategy to selectively capture intact transcripts, which requires RNases generating 5′-hydroxyl and 2′/3′-phosphate termini (e.g., RNase A, RNase I).
• Relies on nonlinear regression software with customisable exponential functions.
Graphical overview
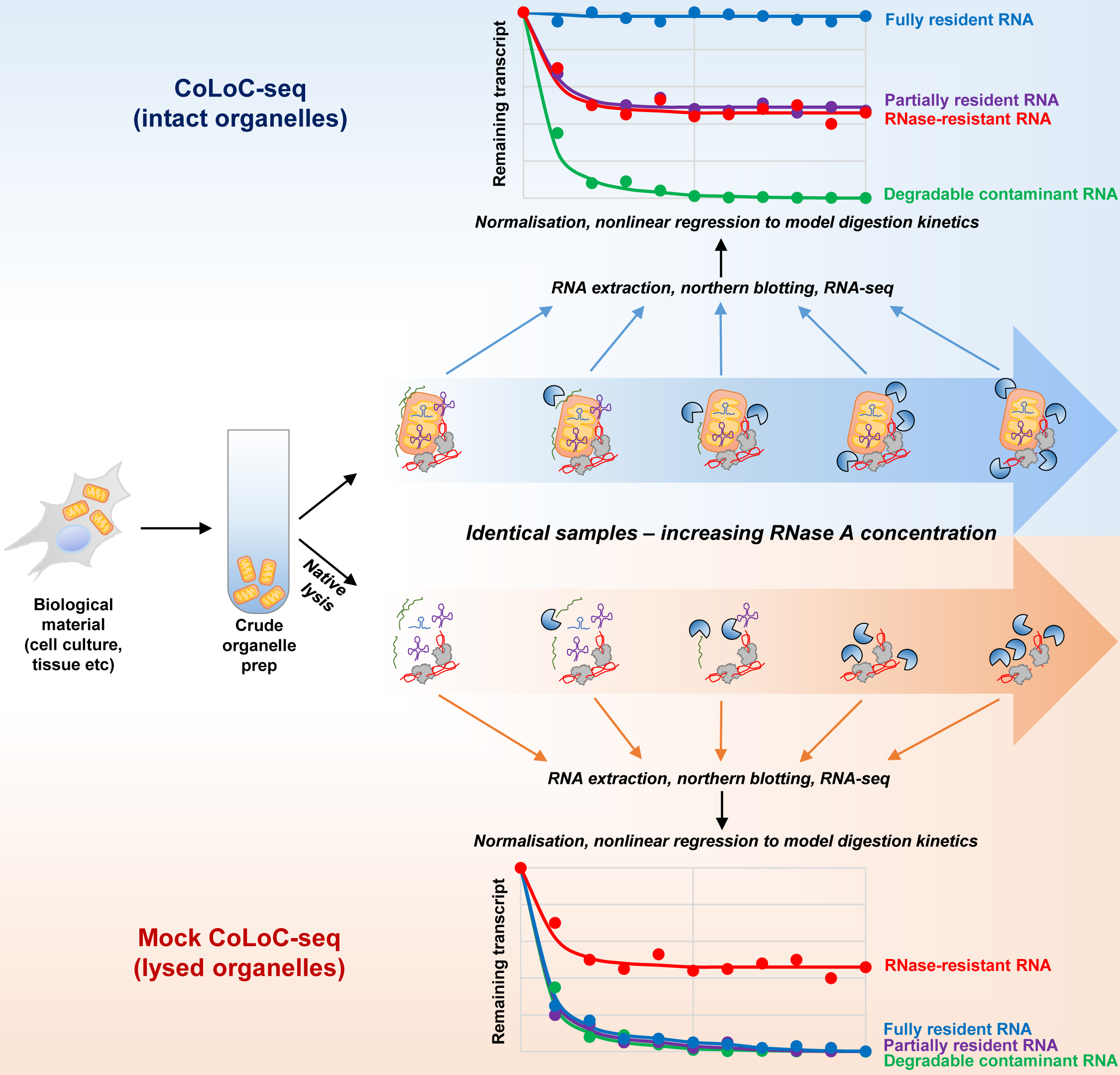
Background
Knowing the localisation topology of transcripts with respect to organelle membranes (inside vs. outside) is critical for the understanding of RNA transactions in various subcellular locations. Selective RNA packaging into viral particles, extracellular vesicles, and ribonucleoproteins (RNPs) attracted much attention over the last two decades (Bresnahan and Shenk, 2000; K. Wang et al., 2010; Arroyo et al., 2011; Routh et al., 2012; Jeppesen et al., 2019; Murillo et al., 2019; Lécrivain and Beckmann, 2020; Gruner and McManus, 2021). Even more intriguing are the intricate interactions between the genetic systems of the nucleus, mitochondria, and plastids inside eukaryotic cells (Woodson and Chory, 2008; L. Levin et al., 2014; Quirós et al., 2016). These organelles possess their own genomes and locally produced transcriptomes. However, in many species, select nuclear-encoded RNAs (primarily tRNAs) enter mitochondria to participate in translation, blurring borders between transcriptomes (Schneider, 2011; Sieber et al., 2011; Jeandard et al., 2019). The scope of such RNA relocation pathways remains insufficiently understood. Therefore, robust genome-wide approaches are required to obtain comprehensive and reliable local transcriptomes.
Confident assignment of RNA localisation topology is challenging, and many approaches have been proposed (Jeandard et al., 2019). In fractionation-based techniques, organelles of interest are purified and treated with a non-specific RNase to degrade contaminant transcripts sticking to their surface. The remaining RNAs, detected by northern blotting, RT-PCR, or RNA-seq, are considered as residing inside the organelles. Although this strategy remains most widely used due to its simplicity and applicability to virtually any species, including non-model, genetically intractable organisms, cell cultures, and tissues (Mercer et al., 2011; Geiger and Dalgaard, 2017), its simple experimental setup turned out to be over-optimistic: some short, structured transcripts embedded in stable RNPs resist degradation, leading to prohibitively high false-positive rates. This caveat is mostly resolved by proximity labelling approaches that use organelle-restricted biochemical tagging of RNA molecules in situ to enable their selective enrichment and identification (Kaewsapsak et al., 2017; Fazal et al., 2019; P. Wang et al., 2019; Zhou et al., 2019; Medina-Munoz et al., 2020; Engel et al., 2021). However, these methods are usually biased against shorter, non-polyadenylated, and lowly abundant transcripts, and require genetic introduction of engineered tagging enzymes targeted to the organelle of interest, which limits their application to model species with tractable genomes and well-characterised protein localisation pathways.
Here, we describe Controlled Level of Contamination coupled to deep sequencing (CoLoC-seq), which marries the accessibility and generality of fractionation-based approaches with the selectivity and robustness of proximity labelling techniques (Jeandard et al., 2023). In a standard CoLoC-seq pipeline (Figure 1, the blue branch), a preparation of intact organelles of interest is split into a suite of samples subjected to a gradient of RNase concentrations. This creates transcript-specific digestion kinetics, amenable to straightforward mathematical modelling, which tells whether a certain RNA fully partakes in the reaction (as expected for contaminants) or if there is a pool of unavailable molecules protected from the RNase. In a parallel Mock CoLoC-seq experiment (Figure 1, the orange branch), the same organelles are first mildly lysed with a detergent to solubilise membranes and then split in a series of identical samples for RNase treatment. Therefore, by measuring the same digestion kinetics in the mildly lysed organelles, one can determine whether the RNA protection is conferred by the organellar membranes or by unrelated factors, such as proteins or intricate structures (such unreactive RNAs are likely false positives).
Successfully tested on human mitochondria, CoLoC-seq outperformed other fractionation-based and proximity labelling approaches, especially in assigning the localisation topology of shorter non-coding RNAs. It can be applied to any RNase-impermeable entity, including eukaryotic organelles, endosymbionts, enveloped and some non-enveloped viruses, and extracellular vesicles.
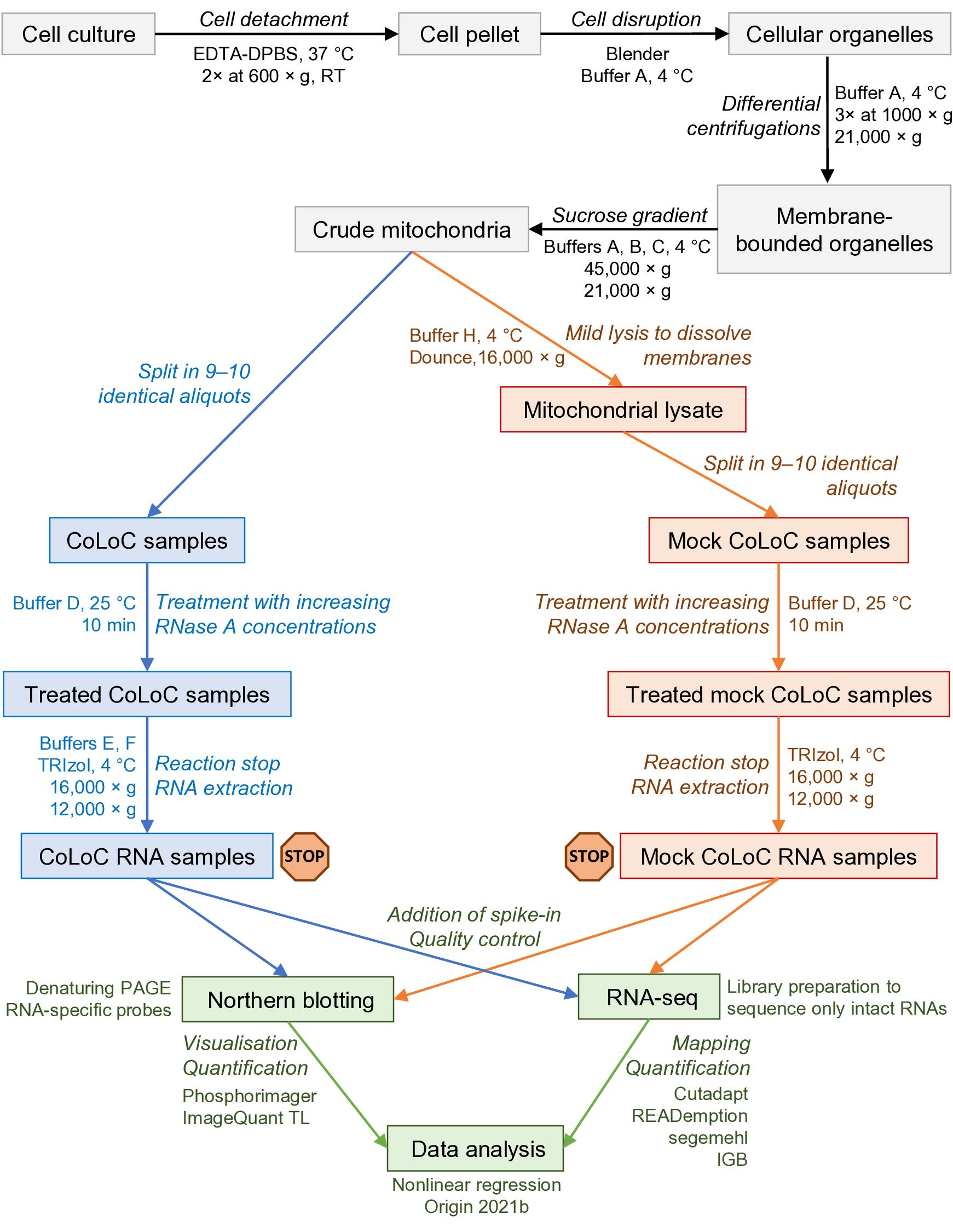
Figure 1. Overview of the protocol and its main steps. The part in grey covers the isolation of crude mitochondria, blue and orange correspond to the CoLoC and Mock CoLoC procedures, respectively, and green is data acquisition and analysis. The stop signs show the steps where the protocol can be safely interrupted without compromising the outcome of the experiment.
Materials and reagents
Biological materials
The CoLoC-seq was performed on mitochondria isolated from the human Flp-In T-REx 293 cells (Thermo Fisher Scientific, catalog number: R78007). One complete set of CoLoC-seq and Mock CoLoC-seq samples requires approximately 2,500 cm2 of nearly confluent cells (equivalent to ~3 × 108 HEK293 cells) devoid of mycoplasma contamination. For application of CoLoC-seq to other systems and cellular compartments, the optimal amount of starting material should be determined empirically.
Reagents
EDTA (Sigma-Aldrich, catalog number: E4884)
Tris base (Sigma-Aldrich, catalog number: 11814273001)
Sucrose (Sigma-Aldrich, catalog number: S0389)
Sorbitol (Sigma-Aldrich, catalog number: S1876)
NaCl (Sigma-Aldrich, catalog number: S9888)
Bovine serum albumin, lyophilized, fatty acid free (Euromedex, catalog number: 1035-70-C)
NaOH (Sigma-Aldrich, catalog number: 655104)
Bromophenol blue (Sigma-Aldrich, catalog number: B0126)
Deionised formamide (Sigma-Aldrich, catalog number: S4117)
Dulbecco’s phosphate buffered saline (DPBS 1×) (Sigma-Aldrich, catalog number: D5773)
20% sodium dodecyl sulfate (SDS) (Euromedex, catalog number: EU-0660-B)
n-dodecyl-β-D-maltoside (Sigma-Aldrich, catalog number: D4641); store at -20 °C
Glycogen (Sigma-Aldrich, catalog number: G8876); store at 4 °C
TBE 10× (Euromedex, catalog number: ET020-C)
Urea (Sigma-Aldrich, catalog number U5378)
40% acrylamide/bis-acrylamide (19:1) (Carl Roth, catalog number: A516.1); store at 4 °C
Ammonium persulfate (Euromedex, catalog number: EU0009-B)
N,N,N’,N’-Tetramethyl ethylenediamine (TEMED) (Euromedex, catalog number: 50406)
Ethidium bromide 1% (Biosolve BV, catalog number: 05412341); store at 4 °C
SSC buffer 20× (Euromedex, catalog number: BI-D0623-1L)
Denhardt’s solution 50× (Thermo Fisher Scientific, catalog number: 750018); store at -20 °C
TE (Tris-EDTA buffer), pH 7.4 (10×) (Euromedex, catalog number: BI-USD8211-1L)
DNase I (Thermo Fisher Scientific, catalog number: EN0525); store at -20 °C
RNase A, DNase- and protease-free, 10 mg/mL (Thermo Fisher Scientific, catalog number: EN0531); store at -20 °C
SUPERase In RNase inhibitor (Thermo Fisher Scientific, catalog number: AM2694); store at -20 °C
AMPure XP kit (Beckman Coulter, catalog number: A63881)
Bradford assay ROTI Nanoquant (Carl Roth, catalog number: K880.1); store at 4 °C
TRIzol reagent (Thermo Fisher Scientific, catalog number: 15596026); store at 4 °C
γ-[32P]-ATP (10 Ci/L, 3,000 Ci/mmol) (PerkinElmer, catalog number: BLU002A100UC). Should be used within approximately one month (32P half-life is 14.268 days); store at -20 °C
Polynucleotide kinase (PNK) and 10× kinase reaction buffer A (Promega, catalog number: M4101); store at -20 °C
Chloroform (Carl Roth, catalog number: 6340.4)
Isopropanol (Carl Roth, catalog number: CP41.1)
Ethanol (Dutscher, Carlo Erba, catalog number: 3086072-CER)
Solutions
Sterile 0.1 M EDTA (see Recipes)
Sterile EDTA-DPBS (see Recipes)
Tris-HCl 0.1 M, pH 6.7 (see Recipes)
Sucrose 3.3 M (see Recipes)
Sorbitol 3 M (see Recipes)
NaCl 1 M (see Recipes)
NaOH 6% (see Recipes)
Buffer A (see Recipes)
Buffer B (see Recipes)
Buffer C (see Recipes)
Buffer D (see Recipes)
Buffer E (see Recipes)
Buffer F (see Recipes)
Buffer H (see Recipes)
RNA loading buffer (see Recipes)
RNA denaturing polyacrylamide gel (see Recipes)
Ammonium persulfate 10% (see Recipes)
Pre-hybridisation buffer (see Recipes)
Hybridisation buffer (see Recipes)
Washing buffer (see Recipes)
Stripping buffer (see Recipes)
Ethanol 80% (see Recipes)
RNase A dilutions in buffer D (see Recipes)
Glycogen 20 μg/μL (see Recipes)
Ethidium bromide 0.0001% (see Recipes)
Recipes
Sterile 0.1 M EDTA (store at 4 °C)
Stir 37.2 g of EDTA in 800 mL of H2O. Add NaOH to adjust pH to 8. Dilute the solution to 1 L with water. Filter solution through a 0.22 μm filter.
Reagent Final concentration Quantity EDTA 0.1 M 37.2 g H2O n/a n/a NaOH n/a n/a Total n/a 1,000 mL Sterile EDTA-DPBS (store at room temperature)
Reagent Final concentration Quantity DPBS (1×) 1× 9.6 g EDTA (0.1 M) 2.5 mM 25 mL H2O n/a 975 mL Total n/a 1,000 mL Tris-HCl 0.1 M, pH 6.7 (store at room temperature)
Stir 12.11 g of Tris base in 800 mL of H2O. Add concentrated HCl under the fume hood to adjust pH to 6.7. Dilute the solution to 1 L with water.
Reagent Final concentration Quantity Tris base 0.1 M 12.11 g HCl (concentrated) n/a n/a H2O n/a n/a Total n/a 1,000 mL Sucrose 3.3 M (store at 4 °C)
Reagent Final concentration Quantity Sucrose 3.3 M 112.86 g H2O n/a n/a Total n/a 100 mL Sorbitol 3 M (sterilise by short autoclaving; store at 4 °C)
Reagent Final concentration Quantity Sorbitol 3 M 54.65 g H2O n/a n/a Total n/a 100 mL NaCl 1 M (store at room temperature)
Reagent Final concentration Quantity NaCl 1 M 5.84 g H2O n/a n/a Total n/a 100 mL NaOH 6% (store at room temperature)
Reagent Final concentration Quantity NaOH 6% 0.6 g H2O n/a 10 mL Total n/a 10 mL Buffer A (store at 4 °C)
Reagent Final concentration Quantity Sorbitol (3 M) 0.6 M 20 mL Tris-HCl (0.1 M, pH 6.7) 10 mM 10 mL H2O n/a 70 mL Total n/a 100 mL Buffer B (store at 4 °C)
Reagent Final concentration Quantity Sucrose (3.3 M) 1.65 M 50 mL Tris-HCl (0.1 M, pH 6.7) 10 mM 10 mL H2O n/a 40 mL Total n/a 100 mL Buffer C (store at 4 °C)
Reagent Final concentration Quantity Sucrose (3.3 M) 0.6 M 18 mL Tris-HCl (0.1 M, pH 6.7) 10 mM 10 mL H2O n/a 72 mL Total n/a 100 mL Buffer D (store at 4 °C)
Reagent Final concentration Quantity Sorbitol (3 M) 0.6 M 20 mL NaCl (1 M) 200 mM 20 mL EDTA (0.1 M) 2 mM 2 mL Tris-HCl (0.1 M, pH 6.7) 10 mM 10 mL H2O n/a 48 mL Total n/a 100 mL Buffer E (store at 4 °C)
Reagent Final concentration Quantity Sorbitol (3 M) 0.6 M 20 mL EDTA (0.1 M) 5 mM 5 mL Tris-HCl (0.1 M, pH 6.7) 10 mM 10 mL H2O n/a 65 mL Total n/a 100 mL Buffer F (store at 4 °C)
Reagent Final concentration Quantity Sorbitol (3 M) 0.6 M 20 mL EDTA (0.1 M) 1 mM 1 mL Tris-HCl (0.1 M, pH 6.7) 10 mM 10 mL H2O n/a 69 mL Total n/a 100 mL Buffer H (store at 4 °C)
Reagent Final concentration Quantity Sorbitol (3 M) 0.6 M 0.2 mL n-dodecyl-β-D-maltoside 1% 10 mg Tris-HCl (0.1 M, pH 6.7) 10 mM 0.1 mL H2O n/a 0.7 mL Total n/a 1 mL RNA loading buffer
Prepare on RNase-free water and add a few crystals of bromophenol blue until the solution has a deep colour but is still transparent; store at -20 °C in 1 mL aliquots.
Reagent Final concentration Quantity SDS (20%) 0.025% 0.0125 mL EDTA (0.1 M) 18 mM 1.8 mL Deionised formamide n/a 8.1875 mL Total n/a 10 mL RNA denaturing polyacrylamide gel
Prepare on RNase-free water. Store at 4 °C and pre-warm before use if urea precipitates.
Reagent Final concentration Quantity TBE 10× 1× 10 mL Urea 8 M 48 g 40% acrylamide/bis-acrylamide (19:1) 6% 15 mL H2O n/a n/a Total n/a 100 mL Ammonium persulfate 10% (store at 4 °C for one month)
Reagent Final concentration Quantity Ammonium persulfate 10% 10 g H2O n/a n/a Total n/a 100 mL Pre-hybridisation buffer (store at room temperature)
Reagent Final concentration Quantity SSC 20× 6× 300 mL Denhardt’s solution 50× 5× 100 mL SDS (20%) 0.2% 10 mL H2O n/a 590 mL Total n/a 1,000 mL Hybridisation buffer (prepare immediately before use)
Reagent Final concentration Quantity Pre-hybridisation buffer 1× 0.45× 9 mL TE 10× 0.5× 1 mL NaCl (1 M) 0.5 M 10 mL Total n/a 20 mL Washing buffer (store at room temperature)
Reagent Final concentration Quantity SSC 20× 5× 250 mL SDS (20%) 0.1% 5 mL H2O n/a 745 mL Total n/a 1,000 mL Stripping buffer (store at room temperature)
Reagent Final concentration Quantity SSC 20× 0.01× 0.5 mL SDS (20%) 0.1% 5 mL H2O n/a 994.5 mL Total n/a 1,000 mL Ethanol 80%
Reagent Final concentration Quantity Ethanol (100%) 80% 800 mL H2O 20% 200 mL Total n/a 1,000 mL RNase A dilutions in buffer D (final volume 200 μL; prepare immediately before use and keep on ice)
To prepare the working RNase A solution at 10 μg/mL, dilute 2 μL of RNase A (10 mg/mL) in 1,998 μL of buffer D and mix well.
Reagent Final concentration Quantity Buffer D volume RNase A working solution (10 μg/mL)
0.1 μg/mL
0.2 μg/mL
0.6 μg/mL
1.2 μg/mL
2.0 μg/mL
2.6 μg/mL
3.2 μg/mL
4.0 μg/mL
6.0 μg/mL
2 μL
4 μL
12 μL
24 μL
40 μL
52 μL
64 μL
80 μL
120 μL
198 μL
196 μL
188 μL
176 μL
160 μL
148 μL
136 μL
120 μL
80 μL
Glycogen 20 μg/μL (aliquot and store at -20 °C)
Reagent Final concentration Quantity Glycogen 20 μg/μL 200 mg H2O n/a 10 mL Total n/a 10 mL Ethidium bromide 0.0001% prepared on RNase-free water (store at 4 °C)
Reagent Final concentration Quantity Ethidium bromide 1% 0.0001% 10 μL RNase-free water n/a 100 mL Total n/a 100 mL
Laboratory supplies
Stericup Quick Release-GV sterile vacuum filtration system 0.22 µm pore size (Merck, catalog number: S2GVU10RE)
Yeast tRNA-derived spike-in transcript, which does not cross-map to the human genome, to enable data normalization:
5′-GAGAAGUAAGCACUGUAAAGGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUGCCUUGUUGGCGCAAUCGGUAGCGCGUAUGACUCUUAAUCAUAAGGUUAGGGGUUCGAGCCCCCUACAGGGCUCCA-3′
Note: Depending on biological material, compatible spike-in transcript(s) should be used, with a sequence that does not cross-map with the genome of the examined organism. A spike-in transcript can be purchased or synthesised by in vitro T7 transcription (as in this case).
Amersham Hybond-N+ membrane optimised for nucleic acid transfer (Cytiva, catalog number: RPN203B)
Single-use spectrophotometer cuvette 1.6 mL Semi-micro type ClearLine (Dutscher, catalog number: 030101)
Micro Bio-Spin P-6 chromatography columns (Bio-Rad, catalog number: 7326200)
Custom DNA oligonucleotide probes for northern blot hybridisation. The oligonucleotides used for CoLoC-seq of human mitochondria are listed in Jeandard et al. (2023), Table S1.
Equipment
Micropipette PIPETMAN P2, 0.2–2 μL (Gilson, catalog number: F144054M)
Micropipette PIPETMAN P20, 2–20 μL (Gilson, catalog number: F144056M)
Micropipette PIPETMAN P200, 20–200 μL (Gilson, catalog number: F144058M)
Micropipette PIPETMAN P1000, 100–1,000 μL (Gilson, catalog number: F144059M)
Vacuum aspiration system (Integra Bioscience VACUSAFE Aspiration System, Fisher Scientific, catalog number: 11636620)
Refrigerated tabletop centrifuge for 50 mL tubes (Eppendorf 5810R, Eppendorf, catalog number: 5811000015)
Refrigerated tabletop centrifuge for 2 mL tubes (Eppendorf 5427R, Eppendorf, catalog number: 5409000010)
Refrigerated high-speed centrifuge (Avanti J-E, Beckman Coulter, catalog number: 369005) with a F0850 fixed-angle aluminium rotor (Beckman Coulter, catalog number: 364640)
Polycarbonate 50 mL bottles with screw cap for the F0850 rotor (Beckman Coulter, catalog number: 357002)
Waring two-speed blender (The Laboratory Store, catalog number: 8010EB)
Refrigerated ultracentrifuge Optima XPN-100 (Beckman Coulter, catalog number: A94469) with a swinging bucket SW 32 Ti (Beckman Coulter, catalog number: 369694)
Open-top thin-wall polypropylene tubes for a swinging bucket SW 32 Ti (Beckman Coulter, catalog number: 326823)
Water bath (e.g., VWR, catalog number: 76308-830)
Dounce homogenizer (VWR, catalog number: 432-0200)
Spectrophotometer (e.g., Eppendorf BioPhotometer 6131, Marshall Scientific, catalog number: E-BP6131)
Spectrophotometer NanoDrop (Thermo Fisher Scientific, NanoDrop 2000, catalog number: ND-2000)
Block heater (e.g., Stuart, DD Biolab, catalog number: 001150)
Gel electrophoresis chamber (e.g., BT Lab Systems, catalog number: BT206)
Low-current power supply (e.g., Consort EV233, Fisher Scientific, catalog number: 10369312)
Gel documentation system (e.g., E.A.S.Y. Doc Plus, Herolab, catalog number: 2809300)
Wet transfer tank (e.g., BT Lab Systems, catalog number: BT306)
High-current power supply (e.g., PowerPac HC, Bio-Rad, catalog number: 1645052)
UV lamp for RNA cross-linking (e.g., Hoefer UVC 500 Ultraviolet Crosslinker, Amersham Life Science, catalog number: 80-6222-50)
Rotating hybridisation oven (e.g., Problot 12 Hybridization Oven, Labnet, catalog number: H1200A-230V)
Plastic sealing machine (e.g., Manual heat sealer SK-SK 210 series, FALC Instruments, catalog number: 638.1430.20)
Phosphorimager plate (e.g., VWR, catalog number: 28-9564-75)
Exposition cassette (e.g., VWR, catalog number: 29-1755-23)
Light eraser for Phosphorimager plates (e.g., InmoClinc Screen X-ray film viewer, MedicalExpo, catalog number: 16300)
Phosphorimager scanner (e.g., GE Typhoon Trio Imager, GMI, SKU: 8149-30-0017)
Portable Geiger counter (e.g., Mini900 Ratemeter, Thermo Scientific, catalog number: MFG017)
Software and datasets
ImageQuant TL (v. 7.0, GE Healthcare)
Cutadapt (version 2.8) (available at https://pypi.org/project/cutadapt/2.8/)
READemption (version 0.4.3) (available at https://reademption.readthedocs.io/en/latest/)
Human genome sequence (Genome Reference Consortium Human Build 38 patch release 13)
READemption uses segemehl version 0.2.0-418 as the read aligner
Data analysis pipeline on Zenodo (https://doi.org/10.5281/zenodo.6389451)
Integrated Genome Browser (v. 9.1.8) (available at https://bioviz.org/)
Origin 2021b (v9.8.5.212, OriginLab Corporation) or similar nonlinear regression software
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Smirnova, A., Jeandard, D. and Smirnov, A. (2023). Controlled Level of Contamination Coupled to Deep Sequencing (CoLoC-seq) Probes the Global Localisation Topology of Organelle Transcriptomes. Bio-protocol 13(18): e4820. DOI: 10.21769/BioProtoc.4820.
分类
分子生物学 > RNA > RNA定位
分子生物学 > RNA > RNA 测序
系统生物学 > 转录组学 > RNA测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













