- EN - English
- CN - 中文
Expression Stability Analysis of Candidate References for Normalization of RT-qPCR Data Using RefSeeker R package
使用 RefSeeker R 软件包对 RT-qPCR 数据标准化的候选参考文献进行表达稳定性分析
(*contributed equally to this work) 发布: 2023年09月05日第13卷第17期 DOI: 10.21769/BioProtoc.4801 浏览次数: 1942
评审: AYŞE NUR PEKTAŞUte Angelika HoffmannHélène Léger
Abstract
When performing expression analysis either for coding RNA (e.g., mRNA) or non-coding RNA (e.g., miRNA), reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) is a widely used method. To normalize these data, one or more stable endogenous references must be identified. RefFinder is an online web-based tool using four almost universally used algorithms for assessing candidate endogenous references—delta-Ct, BestKeeper, geNorm, and Normfinder. However, the online interface is presently cumbersome and time consuming. We developed an R package, RefSeeker, which performs easy and straightforward RefFinder analysis by enabling raw data import and calculation of stability from each of the algorithms and provides data output tools to create graphs and tables. This protocol uses RefSeeker R package for fast and simple RefFinder stability analysis.
Key features
• Perform stability analysis using five algorithms: Normfinder, geNorm, delta-Ct, BestKeeper, and RefFinder.
• Identification of endogenous references for normalization of RT-qPCR data.
• Create publication-ready graphs and tables output.
• Step-by-step guide dialog window for novice R users.
Graphical overview
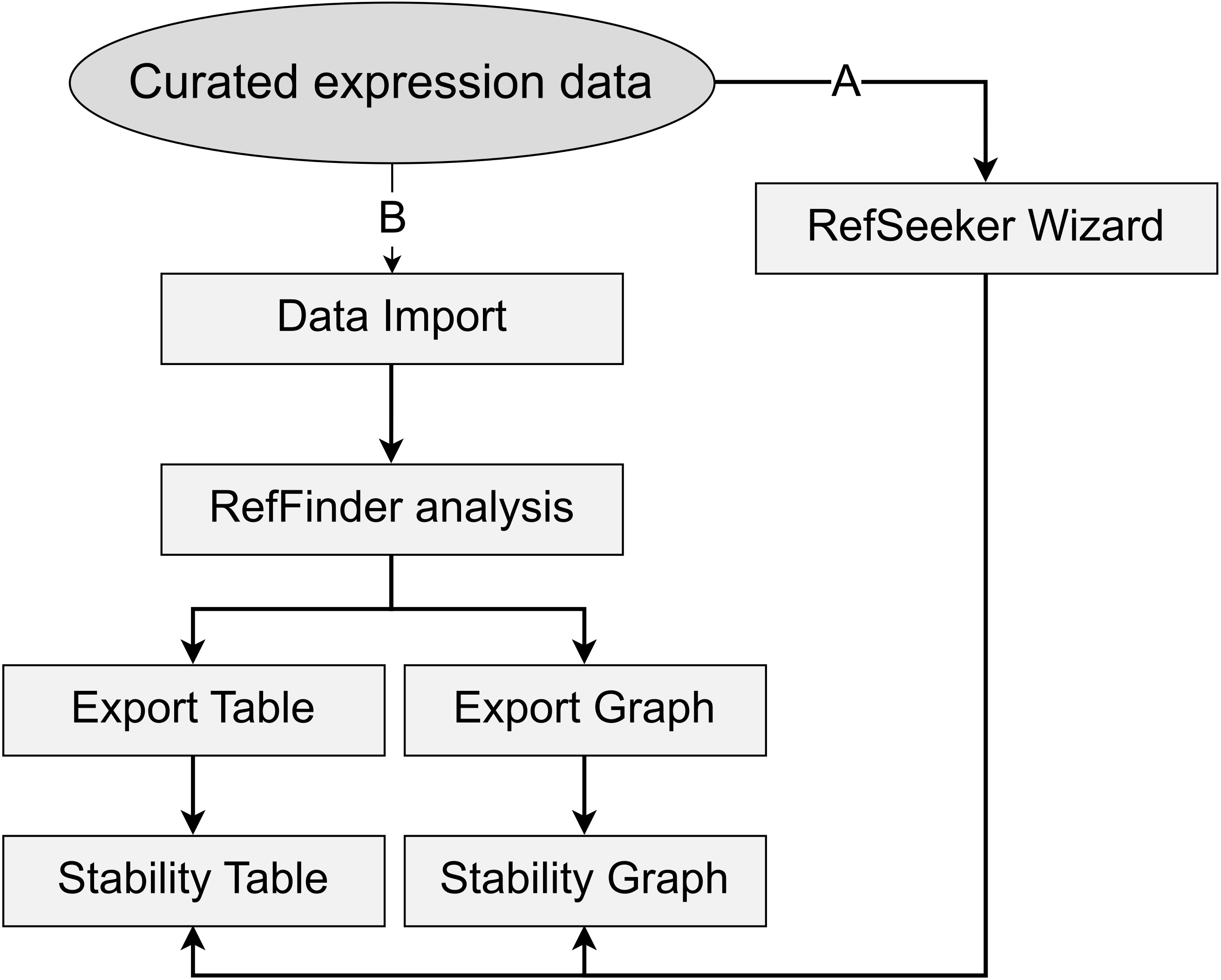
Simple workflow diagram. Two main workflow paths are presented. A) Using the RefSeeker wizard allows non-R programmers to easily load data and choose between selected output formats. B) Command line interface provides more options to control input and output formats and to automate analysis.
Background
Whether coding or non-coding, gene expression research represents a large field of investigation, including molecular biomarker research, drug research, cancer diagnostics, pathway research, RNA interference studies, stem cell research, and much more. In many of these fields, reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) is used to validate results and investigate changes in expression of a variety of RNA types. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guideline requirements have been widely adopted by the scientific community (Bustin et al., 2009). These guidelines assert that the use of three or more stably expressed endogenous references should be used for normalization of target RNAs (e.g., genes/mRNA or miRNAs). Additionally, these references should preferably be of the same type of RNA as the targets.
When performing expression analysis, it is often required to analyze the stabilities of reference genes used to normalize the data from targets of interest (Bustin et al., 2009). The selection of a sufficient number of adequately stable references, typically three or four, is a crucial step since the choice may significantly influence the results and could lead to wrong conclusions (Faraldi et al., 2019). Expression data is often obtained as a set of quantification cycle (Cq), crossing point (Cp), cycle threshold (Ct), or take-off point (TOP) values (Bustin et al., 2009). These expression values are typically obtained by performing the RNA quantification in technical triplicates or quadruplicates, averaging these data results in one value per RNA target (e.g., gene or miRNA) for each sample.
Four different algorithms are commonly used for identifying stable RNAs: (1) Normfinder calculates intra- and inter-group variations (Andersen et al., 2004); (2) geNorm uses an average pairwise standard deviation with all other candidates as the stability measure M (Vandesompele et al., 2002); (3) BestKeeper calculates a range of statistics but bases the individual stability on the mean absolute deviation of the raw Cp values (Pfaffl et al., 2004); and (4) the delta-Ct (ΔCt) method compares each candidate average standard deviation of ΔCt values for each combination of candidates (Silver et al., 2006). Further details on the different algorithms are beyond the scope of this protocol, and further information on their strengths and weaknesses can be found elsewhere (De Spiegelaere et al., 2015; Sundaram et al., 2019).
To deal with differences in results from these algorithms, Xie et. al. created RefFinder, which combines the rankings of the four algorithms and gives a geometric mean of these ranks (Xie et al., 2012 and 2022). RefFinder is available as an online tool, allowing researchers and others to perform the analysis by copying their expression data into a textbox and pressing the analyze button; results are then presented on the webpage.
Given that RefFinder is an online tool, data needs to be copied and pasted into a web-based interface. After the analysis, the results need to be copied and pasted back into a statistical or spreadsheet software of choice for further processing, table generation, and/or graphical depiction. This involves extensive manual work, especially in cases when multiple datasets are used simultaneously or when analyses need to be redone. Moreover, the process can be error prone, considering that copying and pasting manually to and from many different sources and destinations can be disorienting. Therefore, we aimed to develop a straightforward method to perform RefFinder analysis on preprocessed RT-qPCR data, providing easy generation of tables, datasheets, and graphical output: the RefSeeker package.
Software description
RefSeeker is a package developed in R designed to be compatible across different operating systems. R is a widely used, free, and open-source statistical environment, thus providing a great basis for expression data analysis (R Core Team, 2022). R provides great tools for working with data in tabular format as well as for plotting and graphing. The RefSeeker package utilizes widely available tools either available through base R or through The Comprehensive R Archive Network (CRAN).
As for RefFinder, to use the RefSeeker package, expression data need to be prepared in a tabular format. However, data can be prepared either as a data object prepared in R or as one of the supported file types (.xls, .xlsx, .ods, .csv, .tsv, or .txt). Each column represents a named target (gene, miRNA, or other) and each row represents a sample. If the data file is created using R, an index column might be added to the .csv file by default. This should be avoided by setting row.names = FALSE. In case of spreadsheets (Excel or .ods) where more than one dataset is included, each sheet in the spreadsheet file can contain a dataset. Naming the sheets will make it easier to identify the data later, since the name is carried over. In case of txt-based files, e.g., .csv, .tsv, or plain .txt tables, each dataset must be in a separate file in the same folder.
The package functions can be divided into four categories:
1. Data import functions, which import data from different sources (.csv, .tsv, .txt, .xls, .xlsx, and .ods) and arrange it for further processing. A wrapper function, rs_loaddata(), identifies the file extensions and calls the proper import function.
2. Data processing functions. RefSeeker uses four main functions to perform the RefFinder analysis: rs_normfinder(), rs_genorm(), rs_bestkeeper(), and rs_deltact(). These functions are all called by the rs_reffinder() function to determine stability rankings. The comprehensive rank is then calculated as the geometric mean of these stability rankings.
3. Data export functions for further analysis, visualization, and publication of results. The function rs_graph() handles printing and optionally exporting of graphs as .png, .tiff, .jpeg, or .svg file formats. Likewise, the function rs_exporttable() handles export of data tables, either as spreadsheets (.ods or .xlsx), txt-based formats (.csv, .tsv, .txt), or formatted tables in docx format.
4. Interactive implementation of the above functionality through the rs_wizard(). This function provides a graphical user interface dialog window for selecting data and output table and graphical formats.
See Figure 1 for an overview of the main functions and their association to the workflow.
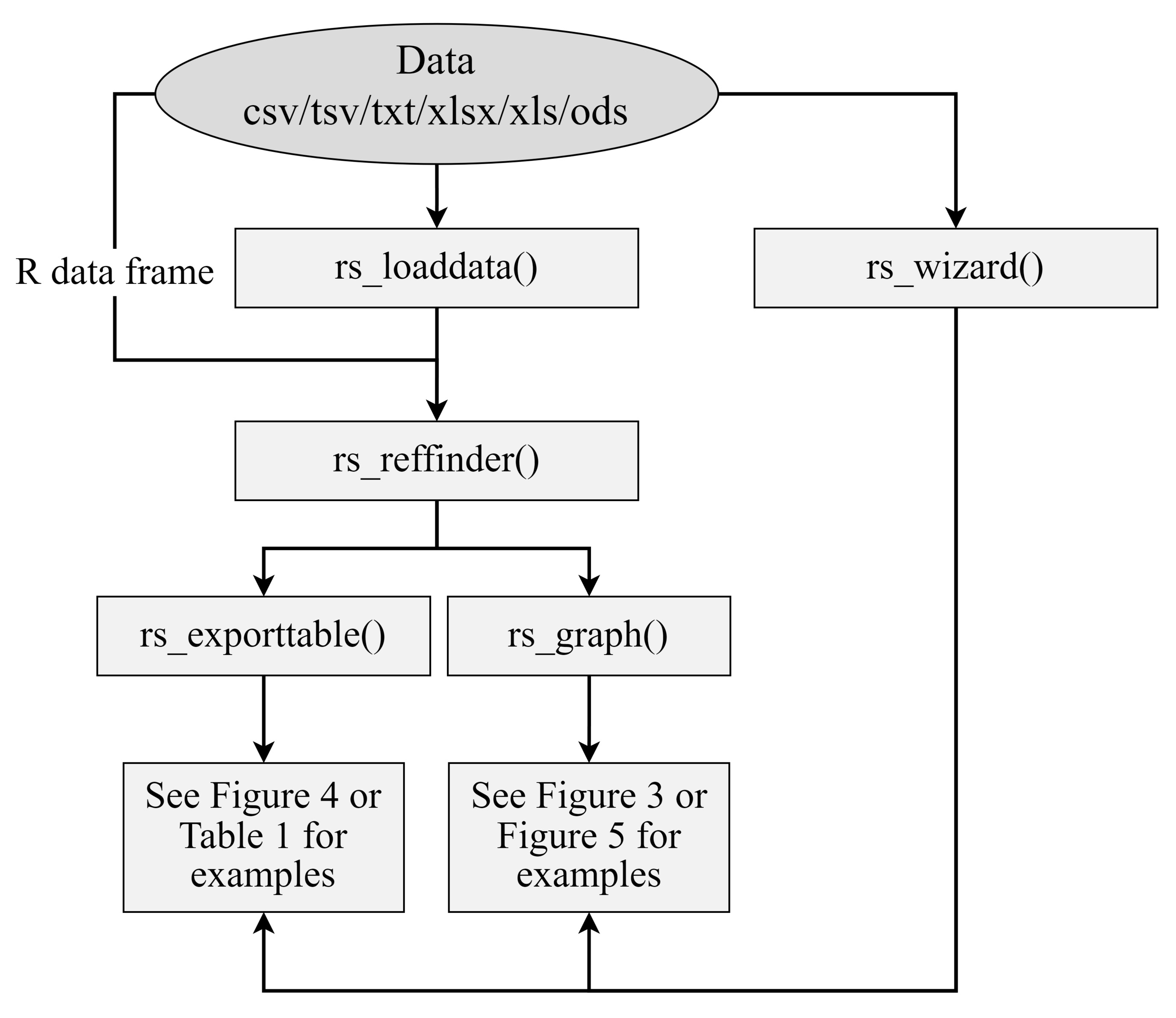
Figure 1. Simple data analysis workflow diagram. Data can be loaded from outside sources via the rs_load() or rs_wizard() functions. RefFinder analysis can be performed on the data using rs_reffinder(), and the analyzed data can be visualized and exported via the rs_exporttable() and rs_graph() functions. Examples of output can be seen in Figure 3, Figure 4, Figure 5, and Table 1.
Equipment
Computer with Windows, MacOS, or Linux-based operating system compatible with R (≥ 4.1.0)
Software and datasets
R software environment (≥ 4.1.0) (https://www.r-project.org/)
RStudio integrated development environment (≥ 1.4.0) (optional, https://rstudio.com/)
Datasets can be prepared in several ways, and processing of raw expression data is outside the scope of this protocol. However, in general, data should be cleaned, quality checked, and adjusted for interplate variability (Petersen et al., 2022).
Data can be prepared in either one of the supported file types (.xlsx, .ods, .csv, .tsv, or .txt) or as a data frame in R.
Sample data can be downloaded from: https://github.com/Hannibal83dk/RefSeeker/blob/main/SampleData/RefSeekerSampleData.xlsx (see Figure 2). These data have been previously described, and details about experiment design, data acquisition, and processing can be found in Petersen et al. (2022), from where the data have been obtained.
• No matter the input source, the following requirements are the same:
• Each column must be representing a gene/target and each row an individual sample*.
• Each column must be named.
• Row names must be excluded.
• No missing data is allowed**.
*Although the RefSeeker package can handle spaces and dashes in column names, some downstream R processes might not be able to. Best practices therefore recommend avoiding these characters in column names.
**Missing data can be handled in several ways. If samples need to be preserved, targets can be removed; if it is more desirable to keep targets, samples can be removed. If both are important, a percentage threshold for allowed missing data can be chosen. This threshold is individually selected; however, it should be as low as possible. A specific recommendation cannot be provided here; however, a threshold of 20% missing data has been used before and seems to be an approximate upper limit. Following target exclusion, remaining missing datapoints can be imputed using different tools [e.g., MissForest (missForrest package), k-Nearest Neighbor (VIM package), Multiple Imputation by Chained Equations (mice package), or max + 1 (manually implemented)].
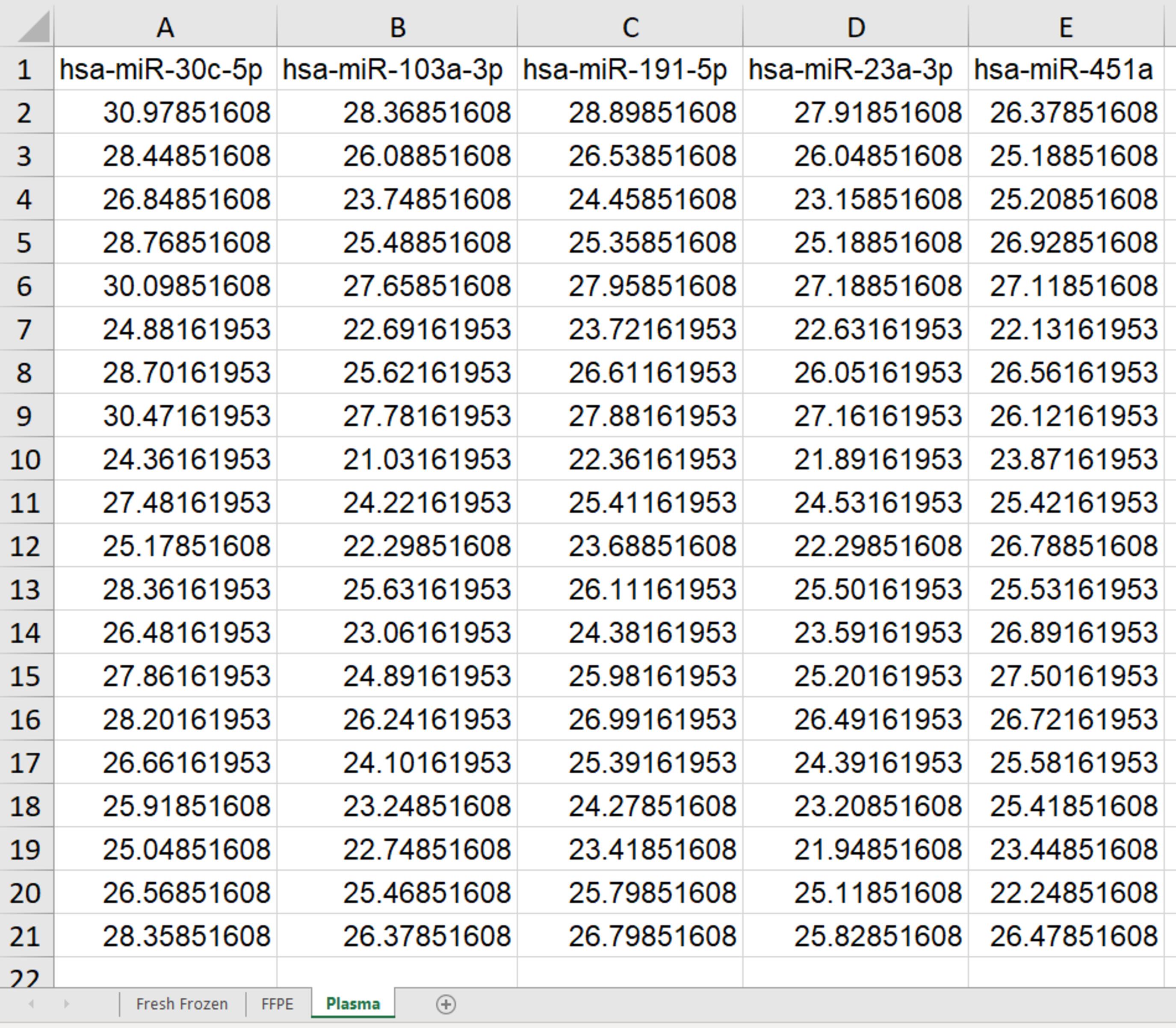
Figure 2. Example of expression data in an Excel file used for RefSeeker analysis. These may be averages of triplicates or quadruplicates and should be adjusted for e.g., possible interplate variation. Targets are given in the first row. Each of the following rows represents raw Cp values (adjusted for interplate variance) obtained from each sample. The file contains three spreadsheets: fresh frozen, formalin fixed and paraffin embedded (FFPE), and Plasma, each containing different datasets.
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Dalsbo Petersen, P. H., Lopacinska-Joergensen, J., Høgdall, C. K. and Høgdall, E. V. (2023). Expression Stability Analysis of Candidate References for Normalization of RT-qPCR Data Using RefSeeker R package. Bio-protocol 13(17): e4801. DOI: 10.21769/BioProtoc.4801.
分类
分子生物学 > RNA > qRT-PCR
生物信息学与计算生物学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link