- EN - English
- CN - 中文
Isoform-specific, Semi-quantitative Determination of Highly Homologous Protein Levels via CRISPR-Cas9-mediated HiBiT Tagging
通过CRISPR-Cas9介导的HiBiT标签对高度同源蛋白水平进行异构体特异性半定量测定
发布: 2023年07月20日第13卷第14期 DOI: 10.21769/BioProtoc.4777 浏览次数: 2521
评审: David PaulManoj B. MenonTalita Diniz Melo Hanchuk
Abstract
Many protein families consist of multiple highly homologous proteins, whether they are encoded by different genes or originating from the same genomic location. Predominance of certain isoforms has been linked to various pathological conditions, such as cancer. Detection and relative quantification of protein isoforms in research are commonly done via immunoblotting, immunohistochemistry, or immunofluorescence, where antibodies against an isoform-specific epitope of particular family members are used. However, isoform-specific antibodies are not always available, making it impossible to decipher isoform-specific protein expression patterns. Here, we describe the insertion of the versatile 11 amino acid HiBiT tag into the genomic location of the protein of interest. This tag was developed and is distributed by Promega (Fitchburg, WI, USA). This protocol describes precise and specific protein expression analysis of highly homologous proteins through expression of the HiBiT tag, enabling protein expression quantification when specific antibodies are missing. Protein expression can be analyzed through traditional methods such as western blotting or immunofluorescence, and also in a luciferase binary reporter system, allowing for reliable and fast relative expression quantification using a plate reader.
Graphical overview
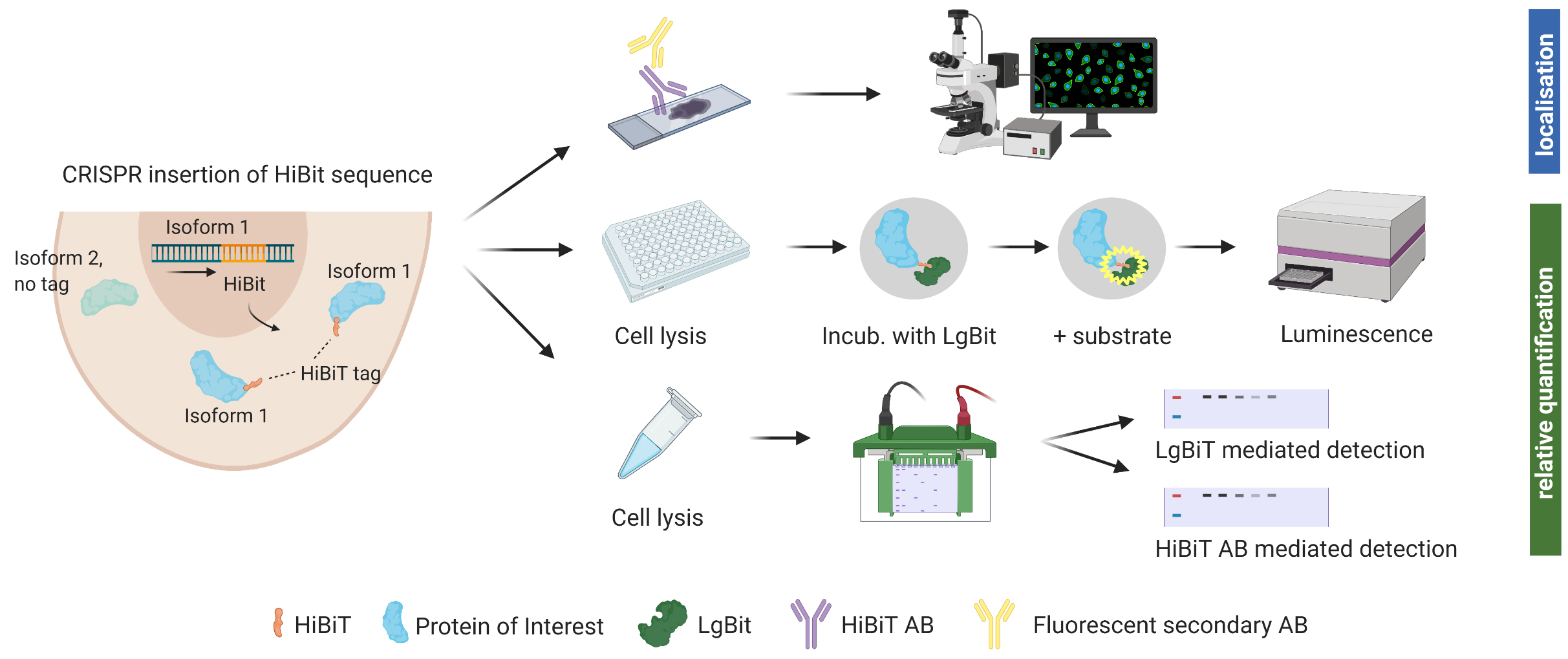
Background
There is hardly a protein to be found within the human body that does not share extensive sequence homology to a family member or isoform, as the human cell requires redundancies to ensure continuous work. Protein isoforms are sometimes encoded by different genes and, often, specific genes have multiple transcripts, returning slightly different proteins depending on how they are spliced. Many diseases are characterized by the predominant expression of specific enzyme isoforms, and isoform expression is often quantified at RNA levels by RT-qPCR, sequencing, or microarrays. Quantifying levels of highly homologous proteins, on the other hand, is difficult and limited by the availability of isoform-specific antibodies. Additionally, batch-to-batch variations in antibody performance as well as recognition of unspecific binding partners sometimes hamper precise protein identification through this method (Voskuil, 2017). Furthermore, when studying newly identified isoforms or lesser studied protein families, there are often no monoclonal, isoform-specific antibodies available. We encountered this issue in our investigation of hexokinase 2 (HK2) and HK3, glycolytic enzymes of the hexokinase family. While sequence homology between HK2 and HK3 is only roughly 54%, we struggled to find appropriate antibodies detecting specifically one isoform or the other when 293T cells were transfected with either HK1, HK2, or HK3 and subsequently stained with a monoclonal antibody for HK2 as well as three antibodies for HK3 (two polyclonal, one monoclonal). The use of polyclonal antibodies should be assessed carefully, as larger immunogens increase the likelihood of producing signals from various family members. If the proteins do not vary in size, their distinction is impossible. HK2 and HK3 are both roughly 100 kDa in size; using antibodies against HK3, we detected signals on western blot in samples that were confirmed HK3-negative through mass spectrometry. This prompted us to generate a protocol for isoform-specific detection of endogenous protein expression without cross-reaction bias.
We hereby suggest the use of an endogenous tag that can be inserted into precise genomic locations and allows relative quantification and identification of protein isoforms through various methods. The system was developed by Promega (Fitchburg, WI, USA) (Schwinn et al., 2018 and 2020). The HiBiT® peptide tag used is 11 amino acids small and binds tightly to an adaptor protein (LgBiT®) when supplied. Through this interaction, the bright luminescent protein NanoBiT® is reconstituted (Dixon et al., 2016). Luminescent intensity from reconstituted NanoBiT is directly proportional to the amount of HiBiT present. This allows for relative protein quantification using a common plate reader. Cell lysate can also be analyzed via SDS gel and subsequent incubation of the membrane with LgBiT®. Furthermore, Promega recently launched an anti-HiBiT monoclonal antibody, which allows visualizing protein localization and expression within a cell using fluorescent microscopy. The anti-HiBiT antibody can also be used in traditional western blotting.
In order to insert the tag into the genomic locus of interest, CRISPR/Cas9-mediated genome editing is used. For successful editing, a sgRNA that binds very close to the desired insertion of the HiBiT tag is complexed with Cas9, and single-strand donor DNA templates (ssODN) for homology-directed repair are supplemented. A few days after electroporation of the cells with Cas9/sgRNA ribonucleoprotein complex (RNP) and ssODN, cells can be assessed for insertion of the tag using a plate reader. While luminescent signal for bulk edited cells in culture was stable during cell propagation, we recommend growing monoclonal populations with 100% of cells expressing the tag for reliable protein quantification.
With the recently launched anti-HiBiT monoclonal antibody, further downstream experiments such as fluorescent microscopy are possible but will require prior optimization.
This example follows the tagging of two hexokinase proteins (HK2 and HK3), two highly homologous proteins encoded on different chromosomes (Figure 1). Here, we show the results of adding the HiBiT tag to the C-terminal end, but inserting the tag at the N-terminus resulted in the generation of a luminescent signal as well. While this strategy is therefore applicable to splice isoforms that differ in either the first or last exon, we do not currently know whether insertion of the tag within the gene body will allow for binding of the adaptor protein and whether this technique can be applied for splice isoforms that share the first and last exon. Insertion of the tag within the gene body could potentially alter protein folding and/or function, presenting a potential limitation for this technique.
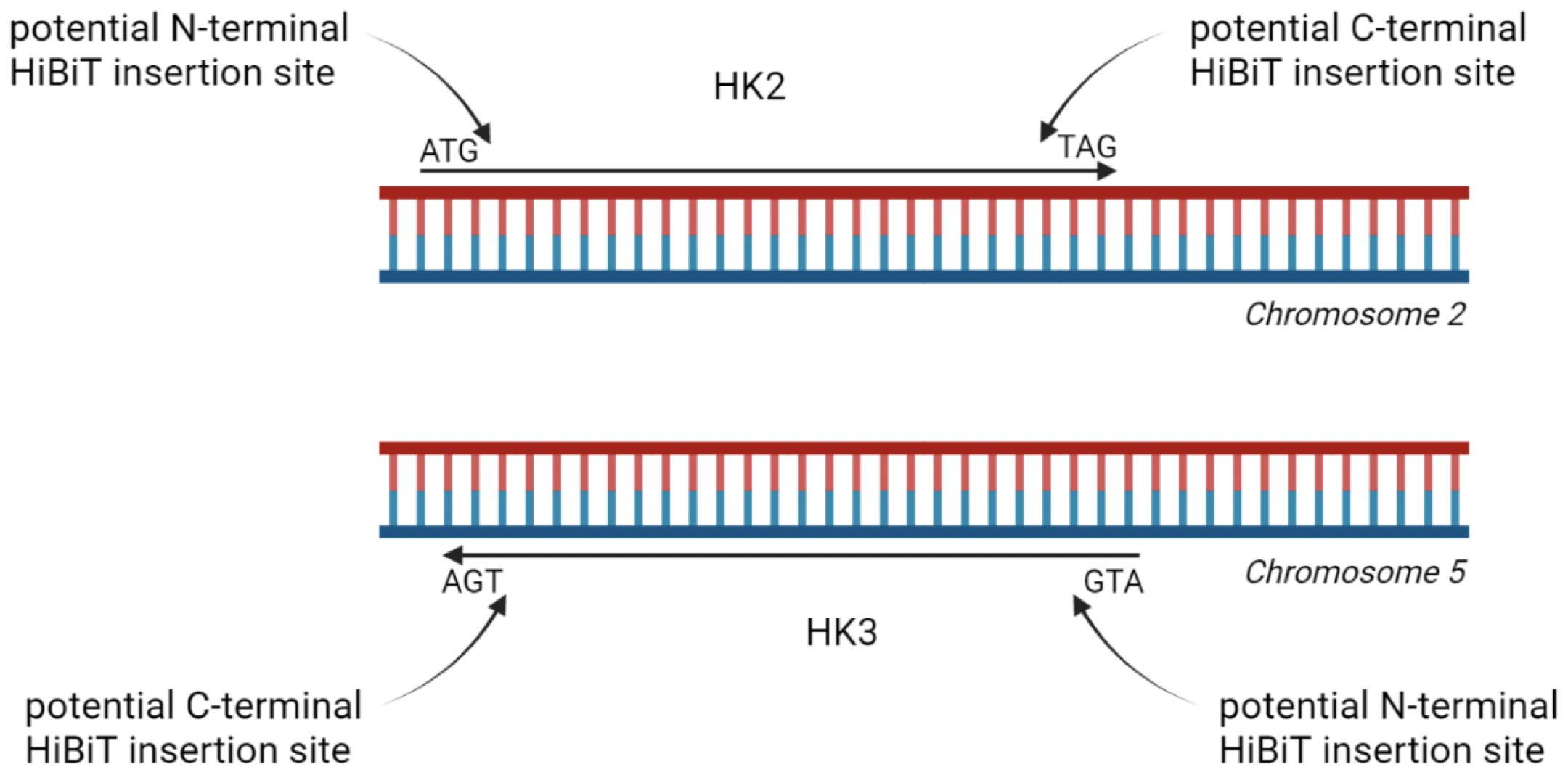
Figure 1. Schematic illustration of tagging options for homologous proteins hexokinase 2 (HK2) and HK3. HiBiT tag can be inserted at either the N- or C-terminal end. Data shown in this protocol are generated with HK2 and HK3 tagged at their C-terminal end.
Materials and reagents
Gene-specific Alt-R® CRISPR-Cas9 sgRNA [Integrated DNA Technologies (IDT)], store at -80 °C
ssODN: gene-specific Alt-R® HDR Donor Oligos (IDT)
eSpCas9-GFP protein (Sigma-Aldrich, catalog number: ECAS9GFPPR-50UG), store at -20 °C
Note: Using a GFP-tagged Cas9 helps visualize electroporation efficiency, but is not needed
Alt-R® HDR enhancer 100 μL (IDT, catalog number: 1081072), store at -20 °C
NeonTM Transfection System 10 μL kit, containing buffers R, T, and E, as well as NeonTM tips and NeonTM electroporation tubes (Thermo Fisher Scientific, catalog number: MPK1025), store buffers at 4 °C after opening; store the remaining components at room temperature (RT)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014)
Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541)
Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S9763)
Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655)
Growth medium suitable for the cell type used [here, we used RPMI 1640 (Sigma Aldrich, catalog number: R8758) supplemented with 10% fetal bovine serum]
Nano-Glo® HiBiT lytic detection system (Promega, catalog number: N3030), store at -20 °C
cOmpleteTM proteinase inhibitor cocktail (Roche, catalog number: 11697498001), store at -20 °C
Nano-Glo® HiBiT blotting system (Promega, catalog number: N2410), store at -20 °C
White microplates suitable for luminescence
4%–20% Mini-PROTEAN® TGC Stain-FreeTM protein gel (Bio-Rad, catalog number: 4568093)
Molecular weight marker, such as Precision Plus ProteinTM KaleidoscopeTM Standard (Bio-Rad, catalog number: 1610375EDU)
Trans-Blot Turbo RTA Mini 0.2 μm Nitrocellulose Transfer kit (Bio-Rad, catalog number: 1704270)
Tris-HCl (Sigma-Aldrich, catalog number: 1083190100)
Triton® X-100 (Sigma-Aldrich, catalog number: X100)
Sodium deoxycholate (Sigma-Aldrich, catalog number: D6750)
Tris base (Trizma® base, Sigma-Aldrich, catalog number: 93352)
Tween® 20 (Sigma-Aldrich, catalog number: P1379)
PBS (see Recipes)
Lysis buffer for HiBiT blotting (see Recipes)
TBS-T (see Recipes)
Equipment
NeonTM transfection system (Thermo Fisher Scientific, catalog number: MPK5000)
Luminescence plate reader (Tecan Group, Tecan Infinite® 200 PRO)
2-D protein electrophoresis equipment (Mini-PROTEAN Tetra Vertical Electrophoresis Cell and PowerPac Power supply, Bio-Rad)
Trans-Blot Turbo transfer system
ChemiDoc XRS+ imaging system (Bio-Rad)
Software
gRNA design tools:
https://chopchop.cbu.uib.no/, https://eu.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE
ImageLab: acquisition of chemiluminescent images using ChemiDoc imaging system
ImageJ: relative quantification of chemiluminescent signal
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Seiler, K., Rafiq, S. and Tschan, M. P. (2023). Isoform-specific, Semi-quantitative Determination of Highly Homologous Protein Levels via CRISPR-Cas9-mediated HiBiT Tagging. Bio-protocol 13(14): e4777. DOI: 10.21769/BioProtoc.4777.
分类
癌症生物学 > 通用技术 > 生物化学试验 > 蛋白质分析
分子生物学 > 蛋白质 > 检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












