- EN - English
- CN - 中文
Lipidomics Workflow for Analyzing Lipid Profiles Using Multiple Reaction Monitoring (MRM) in Liver Homogenate of Mice with Non-alcoholic Steatohepatitis (NASH)
使用多重反应监测(MRM)分析非酒精性脂肪性肝炎(NASH)小鼠肝匀浆中脂质分布的脂质组学工作流程
发布: 2023年07月05日第13卷第13期 DOI: 10.21769/BioProtoc.4773 浏览次数: 2459
评审: Durai SellegounderYu Hui KangSayani DasSibapriya Chaudhuri
Abstract
Non-alcoholic steatohepatitis (NASH) is a condition characterized by inflammation and hepatic injury/fibrosis caused by the accumulation of ectopic fats in the liver. Recent advances in lipidomics have allowed the identification and characterization of lipid species and have revealed signature patterns of various diseases. Here, we describe a lipidomics workflow to assess the lipid profiles of liver homogenates taken from a NASH mouse model. The protocol described below was used to extract and analyze the metabolites from the livers of mice with NASH by liquid chromatography–mass spectrometry (LC-MS); however, it can be applied to other tissue homogenate samples. Using this method, over 1,000 species of lipids from five classes can be analyzed in a single run on the LC-MS. Also, partial elucidation of the identity of neutral lipid (triacylglycerides and diacylglycerides) aliphatic chains can be performed with this simple LC-MS setup.
Key features
• Over 1,000 lipid species (sphingolipids, cholesteryl esters, neutral lipids, phospholipids, fatty acids) are analyzed in one run.
• Analysis of liver lipids in non-alcoholic steatohepatitis (NASH) mouse model.
• Normal-phase chromatography coupled to a triple quadrupole mass spectrometer.
Graphical overview
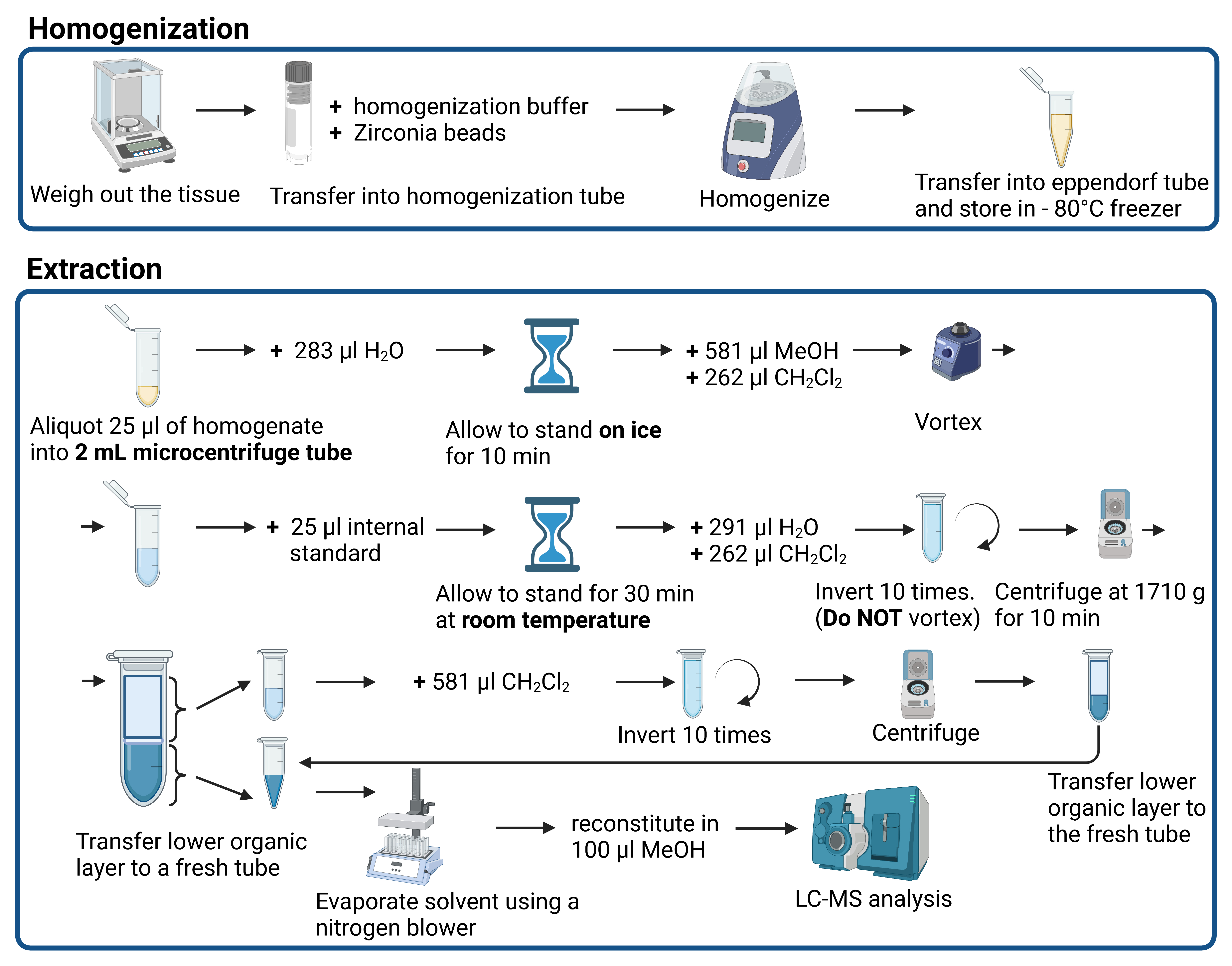
Schematic procedure for the homogenization and extraction of mouse liver tissue in preparation for LC-MS analysis (Created with BioRender.com)
Background
Lipids are essential components of energy metabolism that can be disrupted in metabolic diseases. Lipids comprise various classes, each possessing widely different chemical properties, e.g., steroids are much more non-polar than phospholipids. Due to the widely differing properties, it is difficult to have a single method catered to all the different lipids. As a result, different classes of lipids often have dedicated analytical methods, such as various derivatization techniques, columns, and mass spectrometers, which makes it challenging to have an overview of a broad spectrum of lipids in a single analysis (Wu et al., 2020; Zhao et al., 2020). Another issue of lipid analysis pertains to the triglyceride class. With up to three aliphatic chains attached to a glycerol backbone, the task of identifying and quantifying all species is highly challenging and has been extensively discussed, with each technique having its own limitations and advantages (Han and Ye, 2021). Compared to other methods requiring adaptations, such as two-dimensional LC, argentation, or supercritical fluid chromatography, the method described here provides a broad view of various lipid classes using standard methods that can be done in most laboratories with basic training.
In this protocol, we describe one variant of a lipidomics workflow, originally developed by the mass spectrometry company SCIEX, that covers over five classes of lipids (sphingolipids, phospholipids, glycerolipids, cholesteryl esters, and fatty acids) (Figure 1), including a partial identification of a single aliphatic chain within the neutral lipid, triacylglycerol (Ubhi et al., 2016). The workflow uses a variety of lipid standards, including stable isotopes from ceramides, sphingomyelins, cholesteryl esters, phosphatidylcholines, phosphatidylethanolamines, phosphatidylglycerols, phosphatidylinositols, phosphatidylserines, monoacylglycerols, diacylglycerols, and triacylglycerols. The system uses a normal-phased hydrophilic interaction chromatography (HILIC) column, which separates the lipids by classes as opposed to chain length in reverse-phased systems. In total, the multiple reaction monitoring (MRM) method performs a single-point quantitation of over 1,000 lipid species, providing a convenient overview of common and important lipids for studying metabolic diseases. Here, we apply the techniques to studying the lipidome of liver tissue from a mouse model with non-alcoholic steatohepatitis (NASH). These techniques are not limited to studying the liver but can be applied to other tissues, including cell culture.
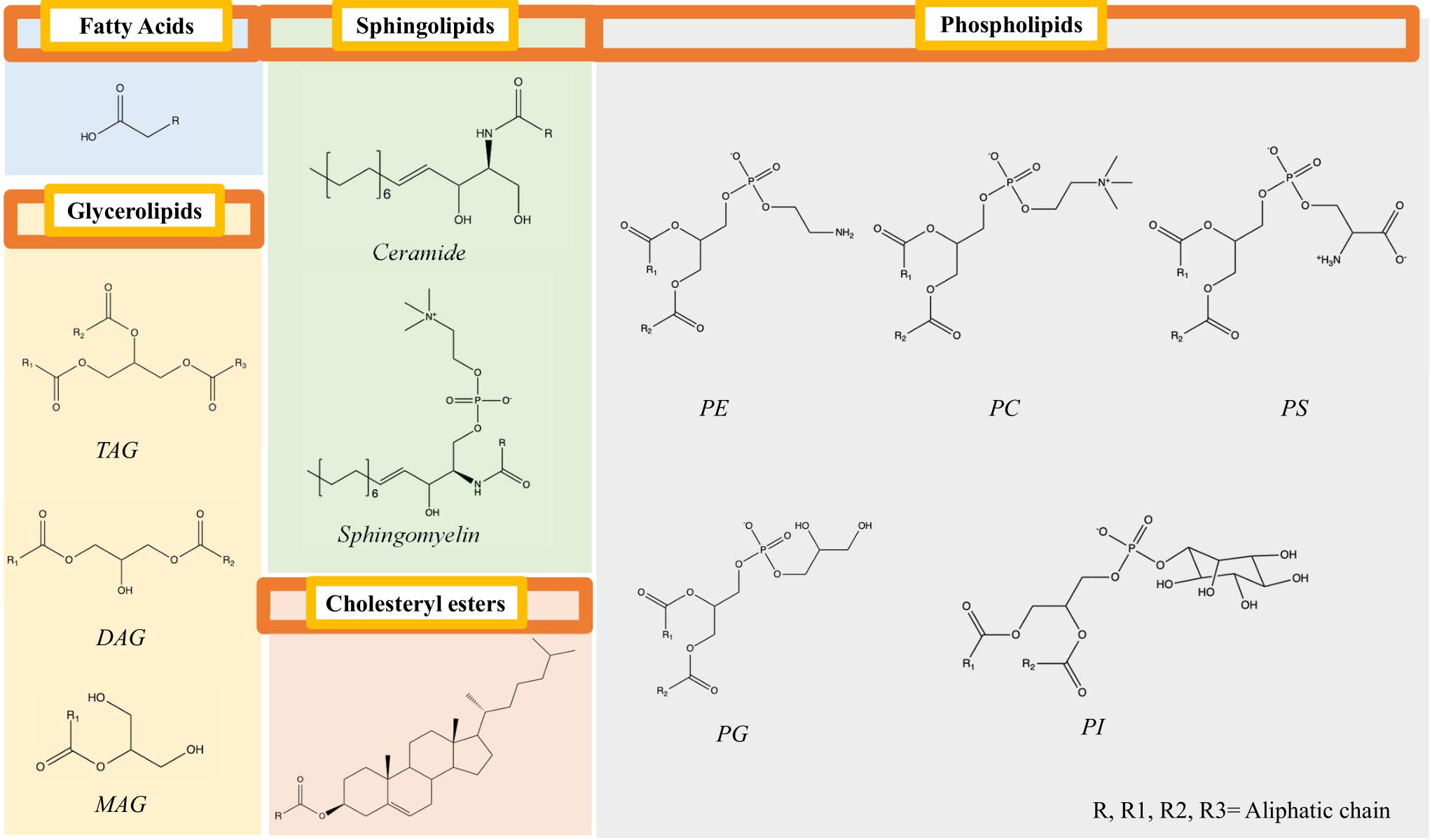
Figure 1. Key structural features of five broad categories of lipids are included in the lipidomics panel
Part I: Tissue homogenization
Materials and reagents
2 mL cryogenic vial (Corning, catalog number: 430488)
2 mL homogenization tubes (Labcon Co., catalog number: 3661-875-000)
Zirconia beads, 1.0 mm diameter (BioSpec Products, catalog number: 11079110ZX)
0.9% sodium chloride solution (Sigma-Aldrich, catalog number: S8776)
Cylinder of liquid nitrogen
Acetonitrile Optima®, LC-MS grade (Fisher chemical, catalog number: A955-4)
Deionized (MilliQ) water (Arium Pro System, Sartorius AG)
Formic acid, HPLC grade (Merck Supelco, catalog number: 5.43804)
Dry ice
Homogenization buffer (see Recipes)
Recipes
Homogenization buffer
Final concentration Amount Acetonitrile n/a 498.5 mL Deionized H2O n/a 498.5 mL Formic acid 0.3% 3 mL
Equipment
Precellys Evolution tissue homogenizer (Bertin Technologies, France)
Cryolys Evolution cooling unit (Bertin Technologies, France)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Wee, H. N., Lee, L. S., Han, S. H. Y., Zhou, J., Yen, P. M. and Ching, J. (2023). Lipidomics Workflow for Analyzing Lipid Profiles Using Multiple Reaction Monitoring (MRM) in Liver Homogenate of Mice with Non-alcoholic Steatohepatitis (NASH). Bio-protocol 13(13): e4773. DOI: 10.21769/BioProtoc.4773.
分类
系统生物学 > 代谢组学 > 脂类组学
生物化学 > 脂质 > 脂质测定
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













