- EN - English
- CN - 中文
Optogenetic Induction of Pyroptosis, Necroptosis, and Apoptosis in Mammalian Cell Lines
光遗传学诱导哺乳动物细胞株的细胞焦亡、细胞坏死性凋亡和细胞凋亡
发布: 2023年07月20日第13卷第14期 DOI: 10.21769/BioProtoc.4762 浏览次数: 3118
评审: Rajesh RanjanTakashi AkeraEmmanuelle BerretAnonymous reviewer(s)

相关实验方案

用于比较人冷冻保存 PBMC 与全血中 JAK/STAT 信号通路的双磷酸化 CyTOF 流程
Ilyssa E. Ramos [...] James M. Cherry
2025年11月20日 2351 阅读
Abstract
Regulated cell death plays a key role in immunity, development, and homeostasis, but is also associated with a number of pathologies such as autoinflammatory and neurodegenerative diseases and cancer. However, despite the extensive mechanistic research of different cell death modalities, the direct comparison of different forms of cell death and their consequences on the cellular and tissue level remain poorly characterized. Comparative studies are hindered by the mechanistic and kinetic differences between cell death modalities, as well as the inability to selectively induce different cell death programs in an individual cell within cell populations or tissues. In this method, we present a protocol for rapid and specific optogenetic activation of three major types of programmed cell death: apoptosis, necroptosis, and pyroptosis, using light-induced forced oligomerization of their major effector proteins (caspases or kinases).
Keywords: Programmed cell death (程序性细胞死亡)Background
Regulated cell death is a common feature of multicellular organisms and plays a key role in development and tissue homeostasis and in protecting the host against malignant growth and various pathogens. Research over the last two decades has identified over 12 different forms of regulated cell death (Galluzzi et al., 2018); however, their study is often complicated by the complex crosstalk and interconnectivity between the different cell death pathways (Bedoui et al., 2020). Additionally, the consequences of different types of cell death in the tissue still remain insufficiently understood, which is at least partially due to the challenges of selectively targeting single cells in multicellular populations, as well as to the pleiotropic effects of commonly used natural cell death triggers both on the dying cells and their neighbors.
In recent years, multiple strategies have been developed to specifically ablate cells both in vitro and in living animals. However, some of these methods (such as laser ablation or photosensitization) still lack specificity regarding the type of cell death to be induced (Tirlapur et al., 2001; Qi et al., 2012), while others [such as chemically inducible dimerization of apoptotic or necroptotic effector proteins (Oberst et al., 2010; Wu et al., 2014)] suffer from a limited spatiotemporal control and require a delivery of soluble ligands, thus limiting their in vivo applications. To overcome these limitations and expand the scope of the tools available for programmed cell death induction, we recently developed a set of optogenetically activated cell death effectors (optoCDEs) (Shkarina et al., 2022), which enable selective induction of three major types of programmed cell death: apoptosis, pyroptosis, and necroptosis. These tools consist of three modules: 1) a photoactuator domain Cry2olig (Cry2 E490G), which responds to blue light by rapid homo-oligomerization, 2) an mCherry tag, which enables the detection of the cells expressing optoCDEs and estimation of the relative construct expression levels, and 3) an effector module (Figure 1A–1C). For opto-caspases, the effector module corresponds to the protease domain (p20 and p10 subunits) of corresponding caspases; the endogenous linkers and cleavage sites essential for the caspase activation are retained, while CARD (in caspase-1, -4, -5, -9, and -11) and DED (caspase-8) domains, responsible for the endogenous upper-level protein–protein interactions and homo-oligomerization, are removed. In opto-RIPK3, the design is similar, while the RHIM motif (responsible for the upstream interaction with the RIPK1) is mutated. In optoMLKL, the effector domain orientation in relation to Cry2olig and mCherry is reversed to keep the MLKL N-terminus (responsible for membrane binding and disruption) exposed. The considerations behind the construct design and testing of the different construct versions are described in more detail in the original paper (Shkarina et al., 2022). The blue light stimulation triggers the rapid activation and oligomerization of Cry2olig, which in turn results in the proximity-induced activation of effector domains and subsequent processing of downstream substrates, culminating in cell death. While Cry2olig alone responds to the blue light within seconds (Taslimi et al., 2014), the timing of cell death induction is defined by the kinetics of effector activation as well as availability and efficiency of the processing and activation of downstream substrates (such as apoptotic executioner caspases, necroptotic effector GSDMD, or pyroptotic effector MLKL); the first morphological features of cell death can usually be detected within minutes after the beginning of illumination.
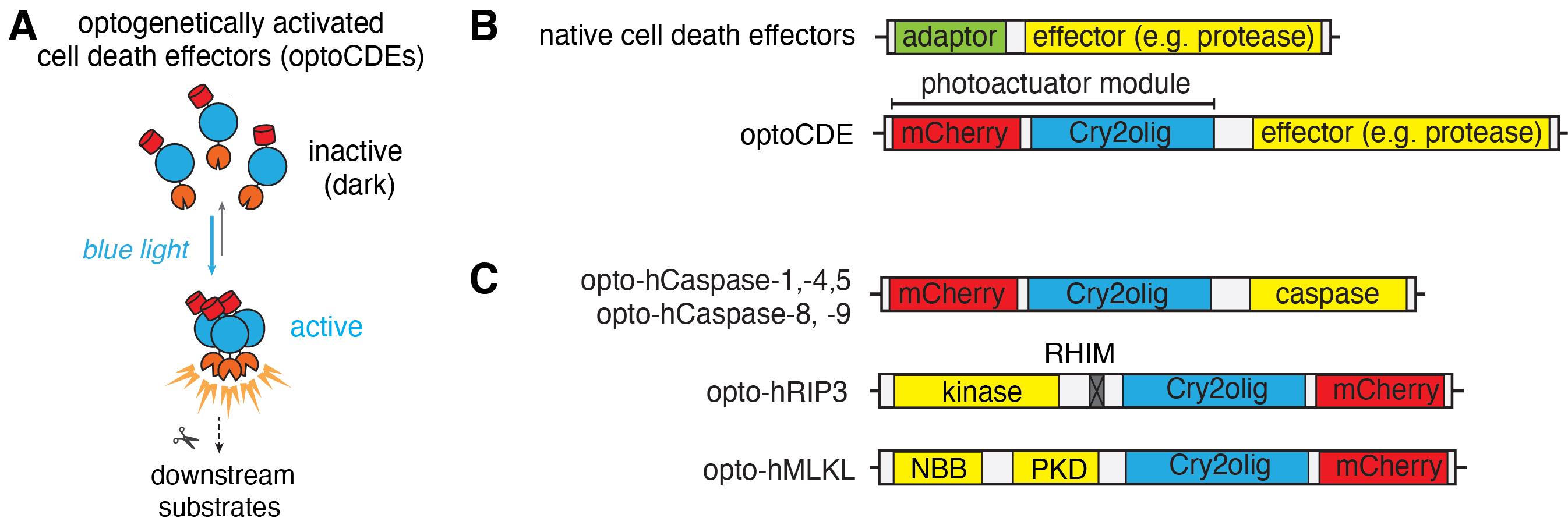
Figure 1. Schematic representation of the optogenetically activated cell death effectors (optoCDE) system. (A) OptoCDEs are inactive and monomeric in the dark state but are activated within seconds upon blue light illumination, which induces oligomerization of the Cry2olig photoactuator domain, activation of downstream substrates, and induction of cell death. (B) General architecture of major cell death effectors and design of optoCDE constructs. Cell death effectors used in the study generally consist of an adaptor domain (CARD or DED for caspases and RHIM for RIPK3) and an effector domain bearing protease (caspases), kinase (RIPK3), or membrane-disrupting (MLKL) function. In the optoCDEs, the effector domain is retained, while the adaptor is replaced with the Cry2olig-mCherry photoactuator module (mCherry is used to visualize optoCDE expression and/or clustering). (C) Schematic representation of major types of optoCDE constructs used in the study. The detailed description and evaluation of the optoCDE tools is available in the original paper (Shkarina et al., 2022). Figure adapted from Shkarina et al. (2022).
The optoCDEs can be applied in vitro, as described in this protocol, as well as in vivo to selectively kill specific cells or cell populations in a highly controlled and specific manner (Shkarina et al., 2022). Additionally, the precise control over the illumination parameters, such as light intensity and duration, provides new means for cellular and mechanistic studies of these forms of cell death, as well as probing cell survival mechanisms that limit cellular damage downstream of these effectors.
Materials and reagents
Cell culture and lentiviral transduction
Sterile serological pipettes (Falcon, catalog number: 357543)
Sterile micropipette tips (Starlab TipOne, catalog number: S1120-8810)
Tissue culture–treated cell culture flasks (TPP, catalog number: 90076)
10 cm Petri dishes (Falcon, catalog number: 351029)
Syringes (5 and 10 mL) (Braun Omnifix, catalog numbers: 4616103V and 4616057V)
Sterile tissue culture–treated 6-well flat-bottom plates (Eppendorf, catalog number: 0030720113)
Sterile 50 and 15 mL Falcon tubes (SPL life sciences, catalog numbers: 50015 and 50050)
Sterile 1.5 mL microcentrifuge tubes (Eppendorf, catalog number: 11.3817.01)
Aluminum foil (Sigma, catalog number: 326852)
0.45 μm filters (Sarstedt, catalog number: 83.1826)
Polybrene (Merck, catalog number: TR-1003-G)
JetPRIME transfection reagent (Polyplus, catalog number: 101000027)
HEPES 1 M (Sigma, catalog number: H3375)
Dulbecco’s phosphate-buffered saline (DPBS) 1× (Thermo Fisher Scientific, catalog number: 10010023)
Dulbecco’s modified Eagle medium (DMEM) with GlutaMAX supplement (Thermo Fisher Scientific, catalog number: 10564011)
RPMI 1640 medium, with GlutaMAX supplement (Thermo Fisher Scientific, catalog number: 61870044)
Doxycycline hydrochloride (Sigma-Aldrich, catalog number: D3447)
Puromycin (Invivogen, catalog number: ant-pr-1)
Hygromycin B Gold (Invivogen, catalog number: ant-hg-1)
LPS-B5 ultrapure (Invivogen, catalog number: tlrl-b5lps)
PMA (Sigma-Aldrich, catalog number: P1585)
Fetal bovine serum (Bioconcept, catalog number: 2-01F10-I)
Live-cell imaging and cell death detection
8-well tissue culture–treated μ slides (ibidi, catalog number: 80826)
Collagen solution from bovine skin (Sigma-Aldrich, catalog number: C4243)
Opti-MEM reduced serum medium (Gibco, catalog number: 11058021)
CellTox Green (Promega, catalog number: G8741)
DRAQ7 (BioLegend, catalog number: 424001)
Annexin V Pacific Blue (BioLegend, catalog number: 640918)
Annexin V FITC (BioLegend, catalog number: 640906)
Annexin V Alexa Fluor 647 (BioLegend, catalog number: 640912)
CellEvent caspase-3/7 green (Thermo Fisher Scientific, catalog number: R37111)
Cell lysis and cytokine secretion analysis
Vision Plate 24, 150 micron, TC-treated, sterile (Life Systems Design, catalog number: 4ti-0241)
96-well flat-bottom plates
Lactate dehydrogenase (LDH) cytotoxicity detection kit (Sigma, catalog number: 11644793001)
Triton X-100 (Sigma, X100-500ML)
Human IL-1β ELISA kit (R&D, catalog number: DY401)
ELISA plates (Sigma-Aldrich, catalog number: M9410-1CS)
LDH stop solution [2 M acetic acid (Sigma, catalog number: 64-19-7)] in dH2O, store at 4 °C
Reagents and equipment for ELISA
Reagents and equipment for the Western Blot analysis
Fluorescence assay (Cisbio, catalog number: 62HIL1BPET)
Equipment
For cell culture
Tissue culture hood (such as HERASAFETM KS, Thermo Scientific)
Cell incubator (FormaTM Steri-CycleTM CO2 Incubator, Thermo Scientific)
Centrifuge (Eppendorf 5810R)
Light microscope (such as Leica DMI6000B)
Pipettes
For imaging
Point-scanning confocal (such as Zeiss LSM800 or Leica SP8)
For cell population–level assays
Light plate apparatus (Gerhardt et al., 2016) equipped with double row of 450 nm light LEDs
Spectrophotometer/ELISA plate reader
Multichannel pipettes
Western blot imager
Software
ZEN (ZEISS, https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html)
Iris (Jeff Tabor laboratory, http://taborlab.github.io/Iris/)
Fiji (NIH, https://imagej.net/software/fiji) (Version 2.3.0)
Software for the plate reader
Microsoft Excel (Microsoft, version 16.69.1)
GraphPad Prism (version 9.3.1.)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Shkarina, K. and Broz, P. (2023). Optogenetic Induction of Pyroptosis, Necroptosis, and Apoptosis in Mammalian Cell Lines. Bio-protocol 13(14): e4762. DOI: 10.21769/BioProtoc.4762.
- Shkarina, K., Hasel de Carvalho, E., Santos, J. C., Ramos, S., Leptin, M. and Broz, P. (2022). Optogenetic activators of apoptosis, necroptosis, and pyroptosis. J. Cell Biol 221(6): e202109038.
分类
细胞生物学 > 细胞信号传导 > 胞内信号传导
生物工程 > 合成生物学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










