- EN - English
- CN - 中文
Quantification of Chromosomal Aberrations in Mammalian Cells
哺乳动物细胞染色体畸变的定量分析
发布: 2023年08月20日第13卷第16期 DOI: 10.21769/BioProtoc.4739 浏览次数: 2833
评审: Khyati Hitesh ShahAmit Kumar DeyAnonymous reviewer(s)
Abstract
Maintenance of genome integrity requires efficient and faithful resolution of DNA breaks and DNA replication obstacles. Dysfunctions in any of the processes orchestrating such resolution can lead to chromosomal instability, which appears as numerical and structural chromosome aberrations. Conventional cytogenetics remains as the golden standard method to detect naturally occurring chromosomal aberrations or those resulting from the treatment with genotoxic drugs. However, the success of cytogenetic studies depends on having high-quality chromosome spreads, which has been proven to be particularly challenging. Moreover, a lack of scoring guidelines and standardized methods for treating cells with genotoxic agents contribute to significant variability amongst different studies. Here, we report a simple and effective method for obtaining well-spread chromosomes from mammalian cells for the analysis of chromosomal aberrations. In this method, cells are (1) arrested in metaphase (when chromosome morphology is clearest), (2) swollen in hypotonic solution, (3) fixed before being dropped onto microscope slides, and (4) stained with DNA dyes to visualize the chromosomes. Metaphase chromosomes are then analyzed using high-resolution microscopy. We also provide examples, representative images, and useful guidelines to facilitate the scoring of the different chromosomal aberrations. This method can be used for the diagnosis of genetic diseases, as well as for cancer studies, by identifying chromosomal defects and providing insight into the cellular processes that influence chromosome integrity.
Graphical overview
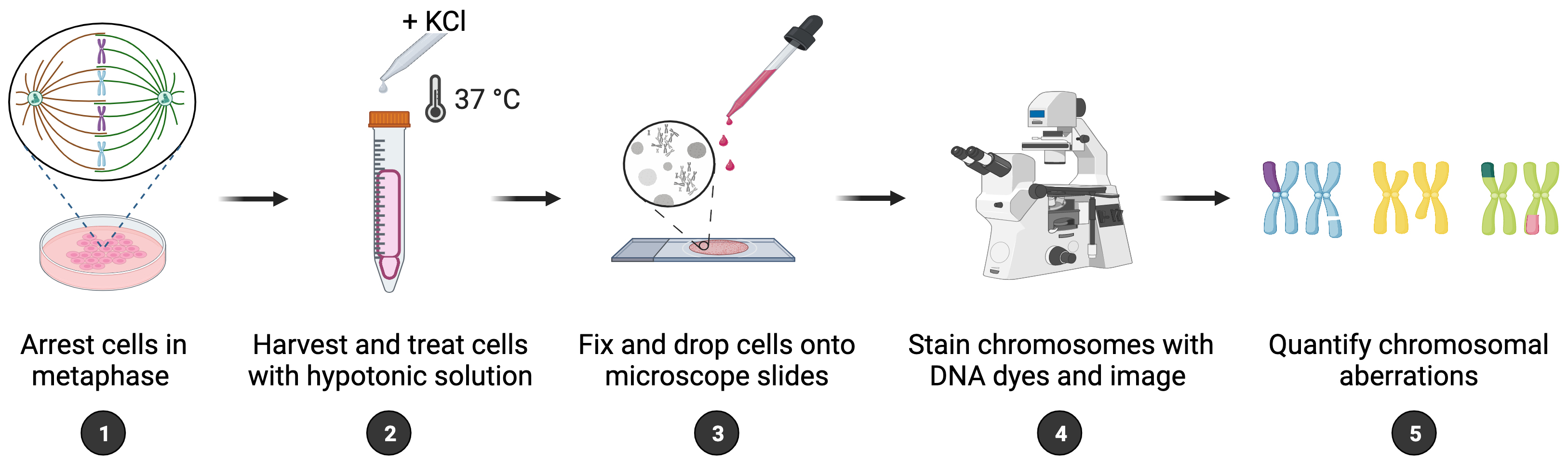
Background
To maintain a stable genome, cells must accurately duplicate their genetic material before each cell division and ensure efficient signaling and repair of DNA damage. Chromosomal instability (CIN) is a form of genomic instability observed in cancer and many congenital abnormalities, typically associated with numerical or structural chromosome changes (Bakhoum and Cantley, 2018). While numerical CIN is characterized by gain and/or loss of whole chromosomes, structural CIN is characterized by gain, loss, and/or rearrangements of parts of chromosomes. Although significant advances in the detection of CIN have been made with the appearance of quantitative high-throughput imaging cytometry and single-cell genomics, classical cytogenetics still remains as an important method to detect chromosomal aberrations within research and clinical settings (Lepage et al., 2019). Cytogenetic approaches generally involve analyzing metaphase chromosomes stained by DNA dyes such as GIEMSA or DAPI; however, inconsistency in the preparation of chromosome spreads is a major problem. Moreover, the scoring of chromosomal aberrations usually relies on specialized experience, since chromosome spreading artifacts can be easily misinterpreted as genomic changes. Thus, efforts should be made to standardize the assays and refine the analysis of chromosomal aberrations for data interpretation. Here, we report a method that enables fast and reliable preparation of metaphase chromosome spreads from mammalian cells for the purpose of scoring chromosomal aberrations. In brief, cells are treated with a metaphase-arresting substance, harvested, and stained with DNA dyes. Metaphase cells are then analyzed microscopically for the presence of chromosomal aberrations. This method can be used to score gross (i.e., observable with standard staining methods) structural aberrations occurring naturally and following exposure to genotoxic chemicals, such as hydroxyurea, aphidicolin, mitomycin C, etc. While numerical aberrations can also be scored, other methods may be more appropriate for that. In addition, we provide representative images of normal and aberrant metaphases as well as scoring guidelines to facilitate accurate identification and evaluation of chromosomal instability, which is of outmost importance within both research and clinical settings.
Materials and reagents
Gloves and lab coat
10 or 15 cm Petri dish (Greiner or Thermo Fisher Scientific, catalog numbers: 664160 or 168381)
15 and 50 mL screw cap tubes (Sarstedt, catalog numbers: 62554502 and 62547254)
Microscope glass slides with a frosted end (Epredia, catalog number: AB00000112EO1MNZ10)
Glass coverslips (24 mm × 50 mm) (VWR, catalog number: 631-1574)
Paper towel
Mammalian cells of interest and appropriate cell culture medium [suggested complete growth medium: Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific, Gibco, catalog number: 41966-029), supplemented with 10% fetal bovine serum (FBS, Capricorn, catalog number: FBS-12A), 100 U/mL penicillin, 100 μg/mL streptomycin (Thermo Fisher Scientific, Gibco, catalog number: 15140-122), and 2 mM L-Glutamine (VWR, catalog number: 392-0441)]
Note: This protocol is for adherent cells.
Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, Gibco, catalog number: 14190144)
Trypsin-EDTA 0.05% (Thermo Fisher Scientific, Gibco, catalog number: 25300-054), store at 4 °C
Fetal bovine serum (FBS) (Capricorn Scientific, suggested catalog number: FBS-12A)
(Optional) Genotoxic chemical e.g., hydroxyurea (Sigma, catalog number: H8627)
Colcemid, KaryoMax colcemid solution, 10 μg/mL (Gibco, catalog number: 15212012), store at 4 °C
Potassium chloride (KCl) (Sigma, catalog number: 1049360500)
Dimethyl sulfoxide (DMSO) (Sigma, catalog number: 34943)
ProLong Gold antifade reagent with DAPI (Thermo Fisher Scientific, catalog number: P36931)
Transparent nail polish (any brand)
Plastic Pasteur pipette (VWR, catalog number: 612-1684P)
Methanol (Honeywell, catalog number: 32213)
Glacial acetic acid (Sigma, catalog number: A6283)
Hydroxyurea (see Recipes)
Hypotonic solution (see Recipes)
Fixative solution (see Recipes)
Equipment
Pipettes (Gilson)
Vortexer
Centrifuge with swing bucket rotor (e.g., Rotina 380, Hettich)
Heating block (Stretching Table OTS 40, Medite, catalog number: 401520)
Water bath (GFL Shaking Water Baths, catalog number: 1083)
Imaging equipment:
Option 1:
Metafer4/MSearch automated metaphase finder system (MetaSystems) equipped with a Zeiss AxioImager Z2 microscope (Carl Zeiss)
Objectives: 10×/0.45 and 63×/1.40 oil Plan-ApoChromat
Filterset: DAPI (Zeiss Filterset 49, ACR)
Camera: CoolCube1
Option 2:
Wide field microscope e.g., Zeiss AxioObserver Z1 microscope
Objective: 63×/1.40 oil Plan-ApoChromat
Filterset: DAPI (Zeiss Filterset 49, ACR)
Camera: ORCA–Flash4.0 V3 Digital CMOS: C13440-20 CU
Software
MetaSystems (version 3.11) software
Fiji (version 2.0) software
GraphPad Prism (version 9) software
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Paniagua, I. and Jacobs, J. J. L. (2023). Quantification of Chromosomal Aberrations in Mammalian Cells. Bio-protocol 13(16): e4739. DOI: 10.21769/BioProtoc.4739.
分类
癌症生物学 > 基因组不稳定性及突变 > 细胞生物学试验 > DNA结构和改变
分子生物学 > DNA > DNA 损伤和修复
分子生物学 > DNA > DNA 结构
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













