- EN - English
- CN - 中文
Biophysical Analysis of Mechanical Signals in Immotile Cilia of Mouse Embryonic Nodes Using Advanced Microscopic Techniques
利用先进显微技术对小鼠胚胎结节中不动纤毛的机械信号进行生物物理分析
发布: 2023年07月20日第13卷第14期 DOI: 10.21769/BioProtoc.4715 浏览次数: 2081
评审: Pilar Villacampa AlcubierreHiroshi YokeChristina Yan Ru Tan
Abstract
Immotile cilia of crown cells at the node of mouse embryos are required for sensing leftward fluid flow that gives rise to the breaking of left-right (L-R) symmetry. The flow-sensing mechanism has long remained elusive, mainly because of difficulties inherent in manipulating and precisely analyzing the cilium. Recent progress in optical microscopy and biophysical analysis has allowed us to study the mechanical signals involving primary cilia. In this study, we used high-resolution imaging with mechanical modeling to assess the membrane tension in a single cilium. Optical tweezers, a technique used to trap sub-micron-sized particles with a highly focused laser beam, allowed us to manipulate individual cilia. Super-resolution microscopy allowed us to discern the precise localization of ciliary proteins. Using this protocol, we provide a method for applying these techniques to cilia in mouse embryonic nodes. This method is widely applicable to the determination of mechanical signals in other cilia.
Keywords: Left-right symmetry breaking (左右对称破坏)Background
Primary cilia—hair-like protrusions on the cell surface—function as antennas. Cilia sense extracellular stimuli, such as flow stimulation, and regulate/organize many signals; one of which governs left-right (L-R) determination (Shinohara and Hamada, 2017). Research concerning flow sensing in cilia began with two techniques: a micropipette (Praetorius and Spring, 2001) and a flow chamber (Nauli et al., 2003) approach. Using these techniques in combination with genetic engineering, we now understand the correlation between flow and chemical reactions, such as calcium responses, within the cilia (Su et al., 2013). However, the sensory capability of cilia with respect to flow, particularly whether cilia sense flow-mediated chemical or mechanical stimuli, is difficult to determine (Ferreira et al., 2019). In this protocol, we provide a method for dissecting the forces acting on cilia.
Recently, Katoh et al. reported the application of optical tweezers (Ashkin, 1970) to a single cilium, which enabled the application of only mechanical force to cilia (Katoh et al., 2018). We further updated this technique to apply to early mouse embryos and combined them with mRNA imaging (Minegishi et al., 2021). We now provide a method for observing the responses to mechanical stimuli by cilia, such as mRNA degradation in ciliated cells (Katoh et al., 2023).
Ascertaining how forces acting on cilia are affected by fluid flow and, in particular, how these forces affect the membrane tension of cilia, is a challenging issue. Owing to the size of cilia, which have a length of ~5 μm and a diameter of only ~200 nm, direct measurement of the tension on the ciliary membrane is difficult. Previously, membrane tension of the cilium was estimated only by flow simulation (Rydholm et al., 2010; Omori et al., 2018). By controlling the extracellular flow and concurrently measuring changes in the shape of cilia using high-resolution microscopy, we first reported the membrane strain of cilia driven by the actual extracellular flow (Katoh et al., 2023).
Finally, we introduced a method for analyzing the precise location of membrane proteins in cilia using 3D stimulated emission depletion (STED) (Klar et al., 2000; Vicidomini et al., 2011), a super-resolution microscope whose lateral resolution is close to 30 nm, using a biophysical approach. In this study, we provide useful protocols for applying state-of-the-art microscopy to cilia in mouse embryos. These methods allow us to study mechanical signals in other cilia as well as in other small organelles.
Materials and reagents
Coverslip (18 mm × 18 mm or 24 mm × 24 mm; No. 1S HT, Matsunami)
400 μm silicone rubber spacer (discontinued product; alternatively, use AS ONE, catalog number: 6-9085-14)
100 μm silicone rubber spacer (AS ONE, catalog number: 6-9085-12)
Rat serum (purchased from Charles River Laboratories Japan)
Fluorescent beads for Point Spread Function (PSF) measurement (Thermo Fisher, catalog number: F8811)
Polystyrene beads for optical tweezers (diameter 3.5 μm) (Thermo Fisher, catalog number: S37224)
Wide bore tip (Funakoshi, catalog number: T-205-WB-C)
FluoroBrite Dulbecco modified Eagle medium (DMEM) (Thermo Fisher, catalog number: A1896701)
Anti-acetylated tubulin antibody (Sigma, catalog number: T6793)
Anti-green fluorescent protein antibody (Abcam, catalog number: ab13970)
Secondary antibody: STAR RED, anti-mouse (Abberior, catalog number: STRED-1001-500UG)
Secondary antibody: STAR ORANGE, anti-chick (Abberior, catalog number: STORANGE-1005-500UG)
2,2’-thiodiethanol (TDE) (Tokyo Chemical Industry, catalog number: T0202)
1,4-diazabicyclo[2.2.2]octane (DABCO) (Sigma-Aldrich, catalog number: 290734-100 ML)
Triton X-100 (Nacalai Tesque, catalog number: 35501-15)
Medium for observation: FluoroBrite DMEM supplemented with 75% rat serum (see Recipes)
PBST (see Recipes)
TDE-DABCO (see Recipes)
Equipment
Microscopy for measurement of flow-dependent changes in ciliary shape (Figure 1A and 1B)
IX83 microscope (OLYMPUS; equipped with shutter (Sh1))
Spinning disk confocal unit (CSU) (Yokogawa, CSU-W1)
Objective lens (Obj.1): UPLAPO100XOHR 1.5 N.A. (Olympus)
UV laser, 375 nm, 70 mW (Kyocera SOC, JUNO 375)
z-Piezo (Physik Instrumente, model: P-721)
FL1: red-light band-pass filter (Asahi, model: LV0630)
Dichroic mirror (DM1) (Chroma, zt405/488/561rpc)
Beam expander (Ex1) (Thorlabs, GBE15-A)
Iris (Linos, G061653000)
Shutter (Sh2) (SURUGA SEIKI, model: F116)
Lens (L1) (Thorlabs, AC254-300-A))
EM-CCD camera (Andor, iXon Ultra 888), water cooled (ASONE, LTB-125A)
Stage top incubator (Tokai Hit, STXF-IX83WX and GM3000)
Optical table (Nippon Boushin Industry, AS-1809)
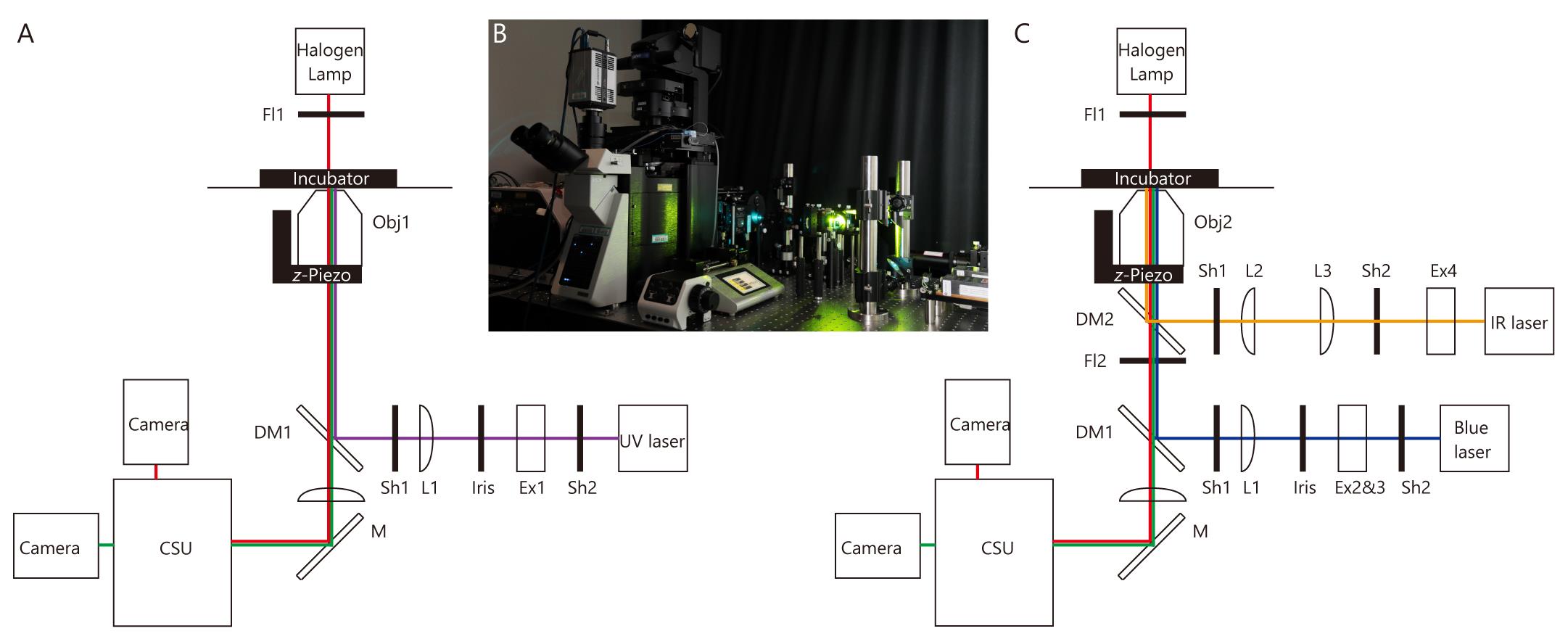
Figure 1. Microscope and optical pathway. (A) Microscope for measurement of flow-dependent changes in ciliary shape. (B) Image of the microscope shown in A and C. Optical pathways are constructed on the optical table. (C) Microscope for optical tweezers and whole-cell fluorescence recovery after photobleaching (FRAP). For abbreviations, see Equipment section in the main text.
Microscope for optical tweezers and whole-cell fluorescence recovery after photobleaching (FRAP) (Figure 1B and 1C)
IX83 microscope (OLYMPUS; equipped with shutter (Sh1))
Spinning disk confocal unit (CSU) (Yokogawa, CSU-W1)
Objective lens (Obj2): UPLSAPO 60XW 1.2 N.A. (Olympus)
z-Piezo (Physik Instrumente, P-721)
Digital analog converter for regulating z-Piezo (USB-DAQ) (National Instruments, USB-6363)
Infrared (IR) laser for optical tweezers, 1,064 nm; 5 W (IPG Photonics, YLR-5-1064-LP-SF)
Blue laser for irradiation, 488 nm; 55 mW (Coherent, Sapphire)
FL1: red-right band-pass filter (Asahi, LV0630)
FL2 (Asahi, SIX870)
DM1 (Chroma, zt405/488/561rpc)
DM2 (Chroma Technology, ZT1064rdc-sp-UF3). Note that we used ZT1064rdc-sp in Katoh et al. (2023), but that DM is better than this.
Beam expander for irradiation (Ex2 and 3) (Sigmakoki, LBED-5 and LBED-3)
Beam expander for optical tweezers (Ex4) (Sigmakoki, LBED-4Y)
Iris (Linos G061653000) (Opened)
Shutter (Sh2) (SURUGA SEIKI, model: F116)
Lens for irradiation (L1) (Thorlabs, AC254-300-A)
Lens for optical tweezers (L2) (Thorlabs, AC254-200-C)
Lens for optical tweezers (L2) (Thorlabs, AC254-150-C)
Neutral density (ND) filter for calibration of optical tweezers (only use for calibration measurement): NENIR13B (Thorlabs; transmission at 1064 nm is 3.81%)
Motorized XY stage (OptoSigma, BIOS-225T and FC-101G)
Stage top incubator (Tokai Hit, STXF-IX83WX and GM3000)
Optical table (Nippon Boushin Industry, AS-1809)
Microscope for analyzing Pkd2 distribution
TCS SP8 STED 3× (Leica)
HC PL APO 100×/1.40 Oil STED white (Leica)
Software
ImageJ, Fiji (version 1.52a, NIH)
Excel (Microsoft)
Software for deconvolution of STED microscopy image (Huygens; version 21.10; Scientific Volume Imaging)
Software for control of IX83 and CSU (iQ; Version 3.6.3; Andor)
Software for control of z-Piezo (LabView 2018; Version 18.0.1f4; National instruments)
Software for analyzing Pkd2 distribution (Igor; version 8.0.4.2; WaveMetrics)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Katoh, T. A., Omori, T., Ishikawa, T., Okada, Y. and Hamada, H. (2023). Biophysical Analysis of Mechanical Signals in Immotile Cilia of Mouse Embryonic Nodes Using Advanced Microscopic Techniques. Bio-protocol 13(14): e4715. DOI: 10.21769/BioProtoc.4715.
- Katoh, T. A., Omori, T., Mizuno, K., Sai, X., Minegishi, K., Ikawa, Y., Nishimura, H., Itabashi, T., Kajikawa, E., Hiver, S., et al. (2023). Immotile cilia mechanically sense the direction of fluid flow for left-right determination. Science 379(6627): 66-71.
分类
细胞生物学 > 细胞运动 > 细胞运动性
发育生物学 > 形态建成 > 能动性
生物物理学 > 单分子技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












