- EN - English
- CN - 中文
Human-rabbit Hybrid Translation System to Explore the Function of Modified Ribosomes
探索修饰核糖体功能的人兔混合翻译系统
(*contributed equally to this work) 发布: 2023年07月05日第13卷第13期 DOI: 10.21769/BioProtoc.4714 浏览次数: 2328
评审: Gal HaimovichFrédéric CATEZVinay PanwarAnonymous reviewer(s)

相关实验方案

小鼠脑裂解物中细胞类型特异性 mRNA 的核糖体翻译亲和纯化 (TRAP)
Catherine L. Salussolia [...] Mustafa Sahin
2022年05月05日 5441 阅读
Abstract
In vitro translation systems are a useful biochemical tool to research translational regulation. Although the preparation of translation-competent cell extracts from mammals has often been a challenge, the commercially available rabbit reticulocyte lysate (RRL) is an exception. However, its valid use, investigating the mechanism of translation machinery such as ribosomes in RRL, presents an analytic hurdle. To overcome this issue, the hybrid translation system, which is based on the supplementation of purified human ribosomes into ribosome-depleted RRL, has been developed. Here, we describe the step-by-step protocol of this system to study translation driven by ribosomes lacking post-translational modifications of the ribosomal protein. Moreover, we combined this approach with a previously developed reporter mRNA to assess the processivity of translation elongation. This protocol could be used to study the potency of heterologous ribosomes.
Keywords: In vitro translation (体外翻译)Background
In addition to transcription, much of gene expression is regulated at the protein synthesis step. Translational regulation is central to diverse cellular and organismal processes, such as development, differentiation, memory, viral infection, tumorigenesis, stress response, and the cell cycle. A wide variety of tools have been developed to monitor the translational output in cells [reviewed in Iwasaki and Ingolia (2017)]. These include fluorescence-based techniques [e.g., fluorescent noncanonical amino acid tagging (Dieterich et al., 2010)], mass spectrometry–based approaches [e.g., pulsed stable isotope labeling by amino acids in cell culture (Schwanhäusser et al., 2009)], ribosome profiling (Ingolia et al., 2009), and single-molecule imaging by Sun tags and spaghetti monster tags (Morisaki et al., 2016; Wang et al., 2016; Wu et al., 2016; Yan et al., 2016). However, for the detailed exploration of molecular mechanisms, in vitro translation remains a powerful and valuable methodology.
Although purified factors could serve for the biochemical reconstitution of protein synthesis processes (Pestova et al., 1998; Shimizu et al., 2001; Pestova and Hellen, 2003; Alkalaeva et al., 2006; Pisarev et al., 2007; Machida et al., 2018; Yokoyama et al., 2019; Abe et al., 2020), the preparation of individual factors is not an easy task. Thus, in vitro translation with cell extracts has become an attractive and useful strategy (Gregorio et al., 2019). For mammals, lysate-based in vitro translation systems were developed with a variety of cell types, such as CHO (Brödel et al., 2014), HEK293 (Fritz et al., 2018), HeLa ( Molla et al., 1991; Bergamini et al., 2000; Witherell, 2001; Thoma et al., 2004; Mikami et al., 2006; Rakotondrafara and Hentze, 2011), and Krebs-2 (Kerr et al., 1966; Mathews and Korner, 1970; Svitkin and Agol, 1978; Svitkin and Sonenberg, 2004 and 2007). Moreover, rabbit reticulocyte lysate (RRL) (Hunt and Jackson, 1974; Pelham and Jackson, 1976; Jackson and Hunt, 1983) has been widely used due to its commercial availability and high translational capacity. However, RRL struggles in depleting proteins by gene knockout or knockdown. Although immunodepletion by antibodies (Rakotondrafara and Hentze, 2011) has been used for this purpose, this technique depends on the efficacy, specificity, and availability of the antibodies for proteins of interest.
This barrier similarly applies to the investigation of ribosomes. Given the growing evidence of modifications of ribosomal RNA (Penzo et al., 2015; Erales et al., 2017; Taoka et al., 2018), ribosomal proteins (Simsek and Barna, 2017), and the heterologous composition of the constituents (Emmott et al., 2019), specialized ribosomes with diverse functions have been proposed (Genuth and Barna, 2018a and 2018b; Guo, 2018). Although ribosome profiling and proteome analysis in vivo provide global views of the function of specialized ribosomes (Ferretti and Karbstein, 2019), biochemical assay systems complement those approaches.
A technique termed the hybrid translation system, which is based on the exchange of ribosomes, is an alternative option in such studies (Panthu et al., 2015; Penzo et al., 2016; Erales et al., 2017; Trainor et al., 2021). Given that ultracentrifugation sediments the ribosome into the pellet, RRL can be prepared as ribosome-free but translation-competent only when the ribosomes are replenished. By supplementation with purified ribosomes of interest, for example from factor-mutated human cell lines, this system makes it possible to test the function of ribosomes in the context of active protein synthesis.
Moreover, the designed reporter enables researchers to investigate the effect of specific codons or RNA sequences along mRNA on translation elongation (Kisly et al., 2018 and 2021). The reporter consists of the fusion of Renilla and firefly luciferases (Rluc and Fluc) and measures the difference in the speeds of Rluc and Fluc synthesis to determine the ribosome elongation rate. Whereas downstream firefly luciferase detection requires complete reporter protein translation (as a whole process of initiation, elongation, and termination), upstream Renilla luciferase can be monitored even in the middle of translation elongation. Thus, the synthesis rate difference between the two luciferases provides a near proxy for elongation efficacy. The insertion of a sequence of interest between the luciferases allows a motif-specific effect on ribosome processivity.
Here, we describe the step-by-step method for the concomitant use of two experimental setups. This protocol includes four major steps: 1) the preparation of ribosome-depleted RRL; 2) the purification of ribosomes from HEK293 cells; 3) the preparation of reporter mRNAs; and 4) the translation reaction. The first two steps were based on an earlier study (Panthu et al., 2015). The design of the reporter mRNA followed the work reported by Tamm and colleagues (Kisly et al., 2021). The combination of the two approaches allowed us to study the effect of the histidine methylation of ribosomal protein uL3 (or RPL3) (Matsuura-Suzuki et al., 2022). Concretely, we delineated the results of ribosomes purified from naïve HEK293T cells and ribosomes purified from cells deficient for METTL18, the enzyme responsible for the methylation of uL3 on His245 (Małecki et al., 2021; Matsuura-Suzuki et al., 2022). In the reporter mRNA, we inserted Tyr repeats between the two luciferases to monitor the ribosome processivity on those codons. Similar applications of this methodology will expand our understanding of the regulatory mechanisms of decorated or specialized ribosomes along diverse codon/RNA element contexts.
Materials and reagents
Pipette, 10 mL, graduated 1/10 mL, sterile, paper-plastic packaging, single packed (Greiner Bio-One, catalog number: 607180)
Pipette, 25 mL, graduated 2/10 mL, sterile, paper-plastic packaging, single packed (Greiner Bio-One, catalog number: 760180)
10 μL, long, graduated, filter tip with system rack (pp), sterilized (WATSON, catalog number: 1252P-207CS)
20 μL hyper filter tip with system rack (pc), sterilized (WATSON, catalog number: 125-20S)
200 μL hyper filter tip with system rack (pc), sterilized (WATSON, catalog number: 125-200S)
1,000 μL, long, graduated, filter tip with system rack (pc), sterilized (WATSON, catalog number: 124-1000S)
TipXL box (IKA Works, catalog number: 0020017832)
Tube, 15 mL, pp, 17/120 mm, natural, sterile, 20 pcs/bag (Greiner Bio-One, catalog number: 188271-013)
Labcon SuperClear 1.5 mL screw cap microcentrifuge tubes (Thermo Fisher Scientific, catalog number: 3611-870-000)
11 mm diameter Delrin tube adapter (Beckman Coulter, catalog number: 393238)
DNA LoBind tubes, 1.5 mL (Eppendorf, catalog number: 0030108418)
Nunc EasYDishes, 100 mm (Thermo Fisher Scientific, catalog number: 150466)
3.2 mL, open-top thick wall polycarbonate tube, 13 mm × 56 mm (Beckman Coulter, catalog number: 362305)
0.2 mL 8-strip PCR tube and cap (NIPPON Genetics, catalog number: FG-028FC)
Screw cap tube, 5 mL (L × Ø): 57 mm × 15.3 mm, PP (SARSTEDT, catalog number: 60.558.001)
Corning 96-well white flat bottom polystyrene not treated microplate, 25 per bag, without lid, nonsterile (Corning, catalog number: 3912)
Rabbit reticulocyte lysate (RRL), nuclease-treated (Promega, catalog number: L4960, stored at -80°C)
DMEM, high glucose, GlutaMAX supplement (Thermo Fisher Scientific, catalog number: 10566016, stored at 4°C)
Fetal bovine serum (FBS) (MERCK, catalog number: F7524, stored at -20 °C)
HEK293T (RIKEN BRC, catalog number: RCB2202)
METTL18 KO HEK293T (Matsuura-Suzuki et al., 2022)
D-PBS(-) without Ca and Mg, liquid (Nacalai Tesque, catalog number: 14249-24, stored at room temperature)
1 M HEPES-KOH buffer solution (pH 7.5) (Nacalai Tesque, catalog number: 15639-84, stored at room temperature)
Potassium acetate (KOAc), nuclease and protease tested (Nacalai Tesque, catalog number: 28434-25, stored at room temperature)
Magnesium acetate tetrahydrate (MgOAc2·4H2O), nuclease and protease tested (Nacalai Tesque, catalog number: 20849-32, stored at room temperature)
Dithiothreitol (DTT), nuclease tested (Nacalai Tesque, catalog number: 14128-62, stored at 4 °C)
UltraPure DNase/RNase-free distilled water (Thermo Fisher Scientific, catalog number: 10977-015, stored at room temperature)
Sucrose, ultra pure (FUJIFILM, catalog number: 198-13525, stored at room temperature)
5 M sodium chloride (NaCl) solution (Nacalai Tesque, catalog number: 06900-14, stored at room temperature)
KCl (2 M), RNase-free (Thermo Fisher Scientific, catalog number: AM9640G, stored at room temperature)
1 M magnesium chloride (MgCl2) solution, sterile filtered (Nacalai Tesque, catalog number: 20942-34, stored at room temperature)
2-Mercaptoethanol, nuclease tested (Nacalai Tesque, catalog number: 21438-82, stored at 4 °C)
Qubit RNA BR Assay kit (Thermo Fisher Scientific, catalog number: Q10210) (accompanied with Qubit RNA BR buffer, Qubit RNA BR reagent, and 0.5 mL PCR tubes)
1 M Tris-HCl solution (pH 6.8) (BioVision, catalog number: 2106-100, stored at room temperature)
UltraPure SDS solution, 10% (Thermo Fisher Scientific, catalog number: 15553-035, stored at room temperature)
Glycerol (Nacalai Tesque, catalog number: 17018-25, stored at room temperature)
Bromophenol blue (Nacalai Tesque, catalog number: 05808-61, stored at room temperature)
BLUE Star prestained protein ladder (NIPPON Genetics, catalog number: MWP03-8, stored at -20 °C)
SuperSep Ace, 5%–20%, 17 well (FUJIFILM, catalog number: 194-15021, stored at 4 °C)
Tris(hydroxymethyl)aminomethane (Nacalai Tesque, catalog number: 35406-91, stored at room temperature)
Glycine (Nacalai Tesque, catalog number: 17109-35, stored at room temperature)
Sodium lauryl sulfate (Nacalai Tesque, catalog number: 31606-75, stored at room temperature)
GelCode blue stain (Thermo Fisher Scientific, catalog number: 24590, stored at room temperature)
Methanol (FUJIFILM, catalog number: 131-01826, stored at room temperature)
Acetic acid (Nacalai Tesque, catalog number: 00212-85, stored at room temperature)
psiCHECK2-Y0× (Matsuura-Suzuki et al., 2022)
psiCHECK2-Y39× (Matsuura-Suzuki et al., 2022)
Primer 1, 5′-TGACTAATACGACTCACTATAGG-3′ dissolved in TE (eurofins, stored at -20 °C) (Matsuura-Suzuki et al., 2022)
Primer 2, 5′-TGTATCTTATCATGTCTGCTCGAA-3′ dissolved in TE (eurofins, stored at -20 °C) (Matsuura-Suzuki et al., 2022)
TE buffer solution (pH 8.0), nuclease and protease tested (Nacalai Tesque, catalog number: 32739-31, stored at room temperature)
PrimeSTAR Max DNA polymerase (TaKaRa, catalog number: R045A, stored at -20 °C)
0.5 M EDTA (pH 8.0) (NIPPON GENE, catalog number: 311-90075, stored at room temperature)
Agarose for ≥ 1 kbp fragment (Nacalai Tesque, catalog number: 01163-05, stored at room temperature)
10× loading buffer (TaKaRa, catalog number: 9157, stored at room temperature)
1 kb DNA Ladder (New England BioLabs, catalog number: N3232S, stored at -20 °C)
GreenView nucleic acid gel stain, 10,000× in water (RELYON, catalog number: N100, stored at 4 °C)
NucleoSpin Gel and PCR Clean-up (MACHEREY-NAGEL, catalog number: 740609.50, stored at room temperature)
T7-Scribe Standard RNA IVT kit (CELLSCRIPT, catalog number: C-AS3107, stored at -20 °C) (accompanied with 10× T7-Scribe transcription buffer, 100 mM ATP, 100 mM CTP, 100 mM UTP, 100 mM DTT, 40 U/μL ScriptGuard RNase inhibitor, T7-Scribe enzyme solution, RNase-free water, and DNase I)
Agencourt RNAClean XP (Beckman Coulter, catalog number: A63987, stored at 4 °C)
Ethanol (99.5) for molecular biology (FUJIFILM, catalog number: 054-07225, stored at room temperature)
ScriptCap m7G capping system (CELLSCRIPT, catalog number: C-SCCE0625, stored at -20 °C)
ScriptCap 2′-O-methyltransferase kit (CELLSCRIPT, catalog number: C-SCMT0625, stored at -20 °C)
A-Plus Poly(A) Polymerase Tailing kit (CELLSCRIPT, catalog number: C-PAP5104H, stored at -20 °C)
RNA 1000 kit (SHIMADZU, catalog number: 292-27913-91, stored at -20 °C and 4 °C), supplied with separation buffer and marker solution
SYBR Green II RNA gel stain, 10,000× concentrate in DMSO (Thermo Fisher Scientific, catalog number: S7564, stored at -80 °C)
RNA 6000 ladder (Thermo Fisher Scientific, catalog number: AM7152, stored at -80 °C)
Formamide (Nacalai Tesque, catalog number: 16229-95, stored at room temperature)
Amino acid mixtures (Promega, catalog number: L4461, stored at -80 °C)
Recombinant RNase inhibitor (TaKaRa, catalog number: 2313A, stored at -20 °C)
Dual-Luciferase Reporter Assay System (Promega, catalog number: E1910, stored at -20 °C), supplied with passive lysis buffer, 5× (Promega, catalog number: E1941, stored at -20 °C)
Liquid nitrogen
DMEM supplemented with FBS (500 mL) (see Recipes)
1 M KOAc (5 mL) (see Recipes)
1 M MgOAc2 (1 mL) (see Recipes)
1 M DTT (5 mL) (see Recipes)
Buffer R (10 mL) (see Recipes)
Sucrose cushion solution (10 mL for eight samples) (see Recipes)
Buffer R2 (5 mL) (see Recipes)
2× Laemmli sample buffer (see Recipes)
10× SDS-PAGE running buffer (see Recipes)
1× SDS-PAGE running buffer (see Recipes)
Gel fixation buffer (see Recipes)
50× TAE (1 L) (see Recipes)
1% agarose gel (100 mL) (see Recipes)
70% ethanol (50 mL) (see Recipes)
Buffer KM (500 μL) (see Recipes)
200 μM amino acid mixture (500 μL) (see Recipes)
1× passive lysis buffer (5 mL) (see Recipes)
Equipment
PIPETMAN P-10 (Gilson, catalog number: F144802)
PIPETMAN P-20 (Gilson, catalog number: F123600)
PIPETMAN P-200 (Gilson, catalog number: F123601)
PIPETMAN P-1000 (Gilson, catalog number: F123602)
PiptPAL single-channel pipette 1,000–10,000 μL (BMBio, catalog number: PAL-10 ml)
Pipet-Aid XP2 110 V, w/Charger (Drummond, catalog number: 4-040-501)
Optima MAX-TL ultracentrifuge (Beckman Coulter, catalog number: A95761)
TLA110 rotor (Beckman Coulter, catalog number: 366735)
Tube rack (13.0 mm, tubes) (Beckman Coulter, catalog number: 348122)
CO2 incubator (PHCbi, model: MCO-170AIC-PJ)
High-speed microcentrifuge (Hitachi, model: himac CF16RN)
Swing rotor (Hitachi, model: T4SS31) and 15TCX6S adaptor (Hitachi, catalog number: S307335A)
High-speed refrigerated microcentrifuge (TOMY, model: MX-307)
High-speed refrigerated microcentrifuge rotar rack (TOMY, model: AR015-24)
Analytical balance (SHIMADZU, model AP124W)
Slim stirrer same rotation control (AS ONE Corporation, model: 1-5940-02-22)
Semimicro stir bar (value) Ø3 mm × 6 mm football (AS ONE Corporation, model: 3-6659-03)
Qubit 2.0 fluorometer (Thermo Fisher Scientific)
Mini cooling dry bath incubator (Major Science, model: MC-0203) with mini dry bath blocks (Major Science, model: MD-MINI-B02)
Power Supply Power Station III (ATTO, model: WSE-3200)
Mini-Gel Slab Electrophoresis Tank (BIO CRAFT, model: BE-211G)
LABO SHAKER (BIO CRAFT, model: BC-740)
Odyssey CLx Imager (LI-COR, model: 9140)
ProFlex 3 × 32-well PCR system (Thermo Fisher Scientific, catalog number: 4484073)
Spectrophotometer (DeNovix, model: DS-11)
Milli-Q reference A+ system (MERCK, model: Z00QSVC01)
Mupid-exU submarine electrophoresis system (ADVANCE, model: EXU-1)
LED transilluminator (Gellex International ltd., model: LB-16)
Ultraviolet transilluminator (UVP, model: M-20)
MIXER uzusio (LMS, model: VTX-3000L)
MINI centrifuge (ALLSHENG, model: Mini-6KS)
NGS MagnaStand (YS-Model) 8 Ch × 0.2 mL PCR tube (FastGene, model: FG-SSMAG2)
Microchip Electrophoresis System for DNA/RNA Analysis MultiNA (SHIMADZU, model: MCE-202), equipped with MICROCHIP, TYPE WE-C (SHIMADZU, model: 292-36010-41)
GloMax Navigator System with Dual Injectors (Promega, model: GM2010)
Software
Image Studio (LI-COR, ver. 5.2)
MultiNA Control Software (SHIMADZU, ver. 1.14.0)
MultiNA Viewer (SHIMADZU, ver. 1.14.0)
GloMax Navigator Software (Promega, ver. 3.1.0)
Excel (Microsoft, ver. 16.66.1)
Procedure
Preparation of ribosome-depleted RRL
Load 1 mL of RRL into a 1.5 mL microcentrifuge tube, place into an 11 mm Delrin tube adapter, and ultracentrifuge at 240,000× g for 2 h 15 min at 4 °C using an Optima MAX-TL ultracentrifuge with a TLA110 rotor.
Note: Keep the sample on ice as much as possible in steps 1–2. Handling the sample in a cold room should be an option.
Collect 900 μL of the supernatant, transfer to a 1.5 mL DNA LoBind tube, flash freeze with liquid nitrogen, and store at -80 °C. See Video 1 for details on the RRL supernatant collection.
Notes:
Avoid touching the precipitate that contains the ribosomes.
Consider dividing the ribosome-depleted RRL into aliquots in several tubes before the flash freezing to avoid repeated freezethaw cycles.
Video 1. Collection of the RRL supernatantPurification of ribosomes from HEK293 cells
Seed 5 × 106 HEK293T (naïve or METTL18 KO) cells in 10 mL of DMEM with 10% FBS (see Recipes) into a 10 cm dish and incubate overnight in a humidified incubator with 5% CO2 at 37 °C. Prepare 10 dishes.
Note: The cell culture typically reaches 70%–80% confluency.
Aspirate the medium from a dish and add 5 mL of ice-cold PBS.
Aspirate the PBS immediately and add 1 mL of ice-cold PBS.
Repeat steps 2–3 for the other nine dishes.
Resuspend cells in the dishes by pipetting, transfer to a 15 mL tube, and centrifuge at 500× g for 3 min at 4 °C, using a refrigerated centrifuge with a swing rotor.
Discard the supernatant and add 1 mL of ice-cold PBS.
Resuspend cells by pipetting, transfer to a 1.5 mL DNA LoBind tube, and centrifuge at 500× g for 3 min at 4 °C, using a refrigerated centrifuge with a fixed angle rotor.
Note: Weigh the 1.5 mL tube before the cell transfer for the next step.
Discard the supernatant and weigh the tube with the cell pellet by the scale.
Notes:
From this step, handle the sample on ice or at 4 °C.
We typically have a ~300 mg cell pellet.
Resuspend the cell pellet in the same volume of buffer R (see Recipes) (e.g., 300 μL of buffer R to 300 mg of cell pellet) and incubate for 15 min on ice.
Note: Buffer R is a low-stringency buffer. Thus, the isolated ribosome may contain the associated factors.
Vortex the mixture for 30 s and centrifuge at 16,000× g for 10 min at 4 °C, using a refrigerated centrifuge with a fixed angle rotor.
Collect the supernatant in a 1.5 mL DNA LoBind tube and mix well. Keep 10 μL of the supernatant in another 1.5 mL DNA LoBind tube for Coomassie Brilliant Blue staining.
Load 300 μL of the supernatant at the bottom of the 3.2 mL polycarbonate tube and then underlay 1 mL of sucrose cushion solution (see Recipes) slowly using a PIPETMAN P-1000 with a long 1 mL tip. See Video 2 for details on the underlaying of sucrose cushion solution.
Notes:
The cell lysate should float on top of the sucrose cushion solution. The interface between the cell lysate and sucrose cushion solution should be clearly visible. If the two solutions are mixed, the ribosome pellet may contain increased contaminants.
Instead of PIPETMAN P-1000 with a long 1 mL tip, a cannula or equivalent can be used.
Video 2. Laying sucrose cushion solution under the cell lysateUltracentrifuge the tube at 240,000× g for 2 h 15 min at 4 °C using an Optima MAX-TL with a TLA110 rotor.
Note: Mark the outside edge of the 3.2 mL polycarbonate tube to indicate the side where the ribosome pellet should be located.
Discard the supernatant by removing thoroughly with a pipette and slowly add 100 μL of buffer R2 (see Recipes) to the bottom of the tube, avoiding disrupting the pellet.
Note: A glassy ribosome pellet should be visible on the outside edge of the tube bottom.
Discard the supernatant immediately and add 30 μL of buffer R2.
Place the tube on a tube rack, add a stir bar to the tube, and then resuspend the solution at 4 °C for 15 min on the magnetic stirrer. See Video 3 for details.
Video 3. Resuspension of the pelleted ribosomes with a stir bar and a magnetic stirrerTransfer the solution to a 1.5 mL DNA LoBind tube. Keep 1 μL of the solution in another 1.5 mL DNA LoBind tube for Coomassie Brilliant Blue staining.
Note: If the pellet does not dissolve completely, stir additionally for 15 min.
Determine the RNA concentration by Qubit RNA BR Assay kit and then determine the ribosome concentration as described below.
Prepare 200 μL of Qubit working solution for each standard and sample by diluting Qubit RNA BR reagent 200-fold with Qubit RNA BR buffer, according to the following table below. When you have one sample to test, prepare [2 (standards) + 1 (sample) + 1] × 200 μL solution.
Reagent Final concentration Amount Qubit RNA BR buffer n/a 800 μL Qubit RNA BR reagent n/a 4 μL Prepare the assay tubes (use 0.5 mL PCR tubes accompanied with Qubit RNA BR Assay kit) according to the following table.
Standards
Reagent Final concentration Amount Qubit working solution n/a 190 μL 20× Standard 1 or 2 (accompanied with Qubit RNA BR Assay kit) 1× 10 μL Samples
Reagent Final concentration Amount Qubit working solution n/a 199 μL Ribosome solution n/a 1 μL Vortex standards and samples for 2–3 s and incubate at room temperature for 2 min.
Turn on Qubit 2.0 fluorometer and select RNA and then RNA Broad Range Assay. Calibrate with the standards and then read the sample.
Select Read Next Sample, followed by Calculate Stock Conc. and then 1 μL to calculate the concentration of RNA in the ribosome solution.
Note: We typically have 1,000–2,000 ng/μL RNA in the ribosome solution.
Convert the RNA concentration to the ribosome concentration. Given the size of rRNAs (18S rRNA, 1.9 kb; 28S rRNA, 5 kb), the formula (number of nucleotides × 320.5) provides a total rRNA molecular weight of 6.9 × 103 × 320.5 = 2.2 × 106. Thus, the molar concentration should be determined as X [ng/μL]/(2.2 × 106). For example, for a ribosome solution with 1,500 ng/μL RNA, the ribosome concentration should be 1,500 [ng/μL]/(2.2 × 106) = 0.68 μM.
Dilute the ribosome solution to 226 nM with buffer R2.
Flash freeze the purified ribosome with liquid nitrogen and store at -80 °C.
Note: Consider dividing the ribosome solution into aliquots in several tubes before the flash freeze to avoid repeated freezethaw cycles.
Determine the protein composition in the ribosome solution by Coomassie Brilliant Blue staining.
Mix the HEK293T cell lysate (from step 11) and ribosome solution (from step 17) with 2× Laemmli sample buffer (see Recipes) in each 1.5 mL DNA LoBind tube according to the table below and denature at 95 °C for 10 min in a dry bath incubator.
Reagent Cell lysate Ribosome solution Sample 10 μL 1 μL Buffer R2 n/a 9 μL 2× Laemmli sample buffer 10 μL 10 μL Load 20 μL of each sample and 5 μL of BlueStar prestained protein ladder into a precast 5%–20% polyacrylamide gel in 1× SDS-PAGE running buffer (see Recipes) set up in the mini-gel electrophoresis tank. Perform electrophoresis at a constant 20 mA for 70 min at room temperature with a power supply.
Fix the gel with 15 mL of gel fixation buffer (see Recipes) for 30 min on a rotary shaker at room temperature and briefly wash the gel twice with Milli-Q water.
Stain the gel with 15 mL of GelCode Blue Safe protein stain for 60 min on a rotary shaker and wash the gel with Milli-Q water.
Destain the gel with Milli-Q water and paper towels until the background signals are reduced.
Note: This typically takes 3 h to overnight.
Visualize the gel on an Odyssey CLx imager and Image Studio with 700 nm channels with the focus at 0.5 mm, which corresponds to the center of the 1 mm thick gel. See Figure 1 for representative protein staining results.
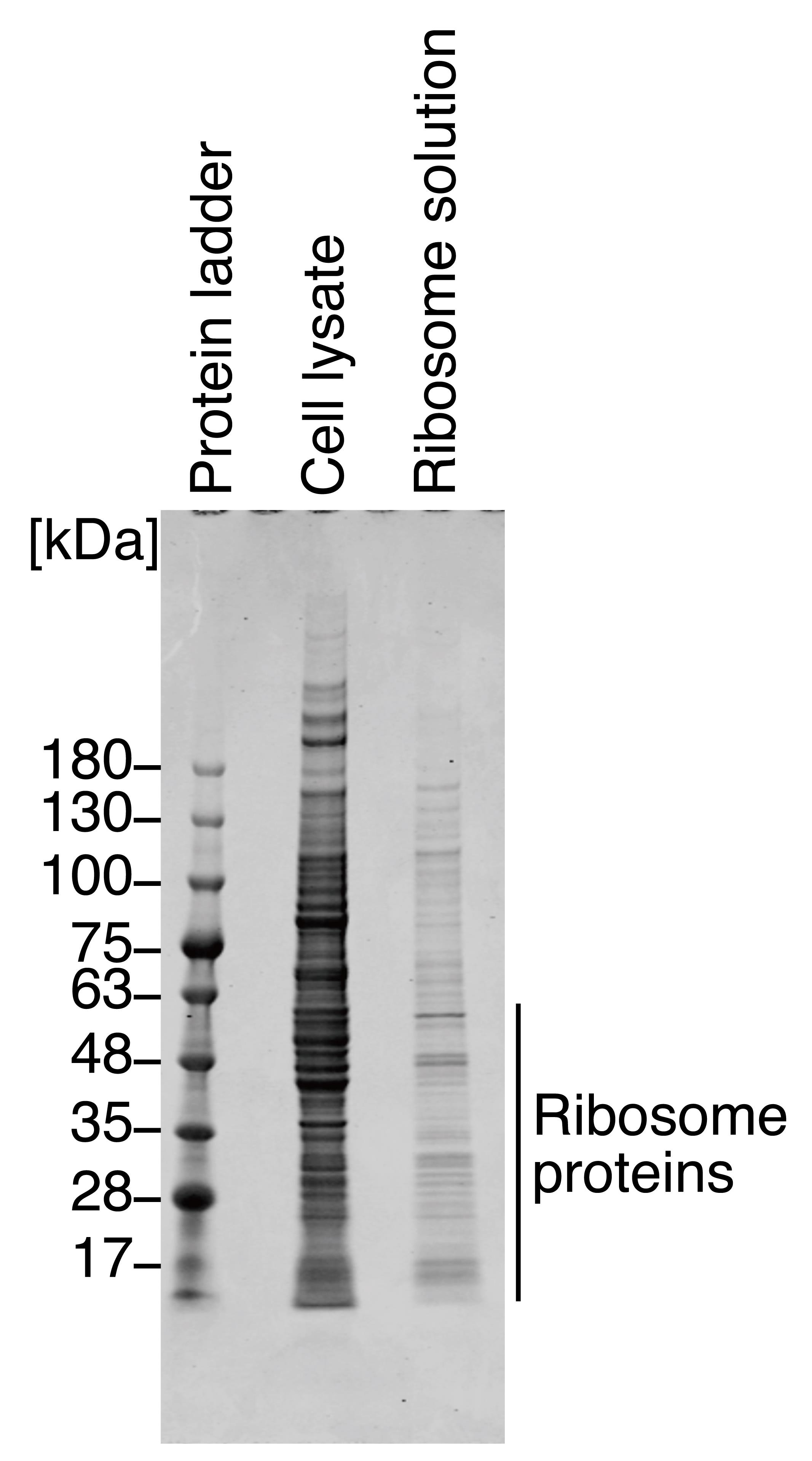
Figure 1. Coomassie Brilliant Blue staining for proteins in ribosome solution. Representative gel image of Coomassie Brilliant Blue staining for proteins in HEK293T cell lysate (i.e., input) and in prepared ribosome solution. The staining pattern was similar to that in an earlier report (Penzo et al., 2016). We note that due to the low stringency of buffer R, the ribosome solution contained high-molecular-weight ribosome-interacting proteins in addition to ribosomal proteins.
Preparation of reporter mRNAs
Prepare the PCR mixture in a 0.2 mL PCR tube as described below to amplify the DNA fragments for the template of in vitro transcription of Rluc-Y39×-Fluc and Rluc-Y0×-Fluc RNAs (100 μL reaction).
Reagent Final concentration Amount 1 ng/μL plasmid (psiCHECK2-Y0× or psiCHECK2-Y39×) 0.01 ng/μL 1 μL 5 μM Primer 1 0.2 μM 4 μL 5 μM Primer 2 0.2 μM 4 μL PrimeSTAR Max Premix (2×) 1× 50 μL RNase-free water n/a 41 μL Perform the PCR amplification in a thermal cycler with the program described below.
Temperature Time Cycles 98 °C 10 s 1× 98 °C 10 s 30× 55 °C 5 s 72 °C 35 s 72 °C 1 min 1× 4 °C ∞ 1× Mix the PCR with 10× loading buffer, load it onto 1% agarose gel (see Recipes) (with 1 kb DNA ladder on a lane), and separate the DNA fragment by electrophoresis with TAE (see Recipes).
Stain the gel with GreenView nucleic acid gel stain in TAE and visualize the nucleic acid using a blue light LED transilluminator. See Figure 2 for the representative results for the Rluc-Y39×-Fluc reporter template.
Gel-purify the appropriate size of DNA fragments (~2.8 kbp) with NucleoSpin Gel and PCR Clean-up according to the manufacturer’s instructions.
Measure the concentration of template DNA using a DS-11 spectrophotometer.
Prepare the in vitro transcription reaction using the T7-Scribe Standard RNA IVT kit in 0.2 mL PCR tubes as described below (20 μL reaction).
Reagent Final concentration Amount Purified DNA template (Rluc-Y39×-Fluc or Rluc-Y0×-Fluc) 1 μg X μL 10× T7-Scribe transcription buffer 1× 2 μL 100 mM ATP 7.5 mM 1.5 μL 100 mM CTP 7.5 mM 1.5 μL 100 mM UTP 7.5 mM 1.5 μL 100 mM GTP 7.5 mM 1.5 μL 100 mM DTT 10 mM 2 μL 40 U/μL ScriptGuard RNase inhibitor 1 U/μL 0.5 μL T7-Scribe enzyme solution n/a 2 μL RNase-free water n/a 7.5 - X μL Incubate at 37 °C for 2 h in a thermal cycler.
Note: Extension of the incubation time to 4–6 h may increase the yield of RNA.
Add 1 μL of DNase I (a component of the T7-Scribe Standard RNA IVT kit) and incubate at 37 °C for 2 h in a thermal cycler.
Note: RNA may be stored at -20 °C before purification.
Purify the RNA as described below.
Mix the bottle of the Agencourt RNAClean XP thoroughly to achieve homogeneous resuspension.
Add 1.8× volume of bead solution to the reaction, mix well by pipetting 10 times or vortexing for 30 s, and centrifuge the tube briefly.
Incubate at room temperature for 15 min and place the tube on the magnetic stand for 5 min to separate the beads from the solution.
Discard the cleared supernatant.
Add 200 μL of 70% ethanol (see Recipes) into the tube and incubate for 30 s at room temperature.
Note: Keep the tube standing on the magnetic stand during steps e–f.
Discard the cleared supernatant.
Repeat steps e–f twice (for a total of three washes).
Centrifuge the tube briefly to collect the remaining 70% ethanol at the bottom of the tube, keep the tube on the magnetic stand for 1 min, and discard the cleared supernatant.
Open the lid of the tube and allow the beads to dry for 5 min at room temperature.
Note: Beads may be cracked after completely drying.
Caution: Overdrying the beads may result in inefficient recovery of RNA.
Add 20 μL of RNase-free water, resuspend the beads by pipetting, and incubate for 2 min at room temperature.
Place the tube on the magnetic stand for 2 min at room temperature and then transfer the cleared supernatant to a new 1.5 mL DNA LoBind tube.
Note: The RNA may be stored at -20 °C or -80 °C.
Measure the concentration of the RNA using a DS-11 spectrophotometer.
Note: We typically have ~1 μg/μL RNA solution.
Prepare the following solution in 0.2 mL PCR tubes (33.5 μL reaction volume).
Reagent Final concentration Amount Purified RNA 20–30 μg X μL RNase-free water n/a 33.5 - X μL Note: Keep the remaining RNA for application to the fragment analyzer.
Incubate at 65 °C for 5 min in a thermal cycler and then immediately place on ice.
Prepare the capping reaction using the ScriptCap m7G Capping System and ScriptCap 2′-O-Methyltransferase kit in the 0.2 mL PCR tubes as described below (50 μL reaction).
Reagent Final concentration Amount Denatured RNA 20–30 μg 33.5 μL 10× ScriptCap capping buffer 1× 5 μL 10 mM GTP 1 mM 5 μL 20 mM SAM 0.5 mM 1.25 μL 40 U/μL ScriptGuard RNase inhibitor 1 mM 1.25 μL 100 U/μL ScriptCap 2′-O-methyltransferase 4 U/μL 2 μL 10 U/μL ScriptCap capping enzyme 0.4 U/μL 2 μL Incubate at 37 °C for 30 min in a thermal cycler.
Prepare the poly(A) tailing reaction using the A-Plus Poly(A) Polymerase Tailing Kit in 0.2 mL PCR tubes as described below (66 μL reaction volume).
Reagent Final concentration Amount Capping reaction n/a 50 μL 40 U/μL ScriptGuard RNase inhibitor 0.18 U/μL 0.3 μL 10× A-Plus poly(A) tailing buffer 1× 6.6 μL 20 mM ATP 2 mM 6.6 μL 4 U/μL A-Plus poly(A) polymerase 0.15 U/μL 2.5 μL Incubate at 37 °C for 30 min in a thermal cycler.
Add 2.5 μL of 0.5 M EDTA to stop the poly(A) tailing reaction (to make ~18 mM EDTA at final concentration).
Note: The poly(A) tailing reaction may be stored at -20 °C before purification.
Purify the RNA as described in step 10 above.
Note: Purified RNA may be stored at -20 °C or -80 °C before use.
Assess the purity of the RNA with the fragment analyzer MultiNA.
Note: Instead of MultiNA, Bioanalyzer, TapeStation, or electrophoresis with denaturing TBE-agarose gel can be used.
Bring separation buffer (a reagent of the RNA 1000 kit), marker solution (a reagent of the RNA 1000 kit), SYBR Green stock solution, and RNA 6000 ladder to room temperature.
Dilute SYBR Green stock solution 100-fold with TE.
Dilute RNA 6000 Ladder 6-fold with TE.
Dilute poly(A)-tailed reporter RNA (from step 18) and non-poly(A)-tailed RNA (from steps 10–11) with RNase-free to 25–250 ng/μL.
Prepare the required volume of buffer solution for MultiNA into a screw cap tube, 5 mL, as described below.
Note: The required volume depends on the sample number. The following table shows an example.
Reagent Final concentration Amount 1/100 diluted SYBR Green n/a 1 μL Formamide n/a 80 μL Separation buffer (a reagent of the RNA 1000 kit) n/a 319 μL Prepare the sample and RNA 6000 ladder in 0.2 mL PCR tubes for the MultiNA run as described below.
Reagent Final concentration Amount 1/6 RNA 6000 ladder, 1/10 poly(A)-tailed reporter RNA, or 1/10 non-poly(A)-tailed reporter RNA n/a 3 μL RNA marker solution (a reagent of the RNA 1000 kit) n/a 3 μL Incubate at 65 °C for 5 min in a thermal cycler and immediately place on ice.
Set the 0.2-mL PCR tubes from step f and the 5 mL tube from step e (leaving the lid off) on MultiNA and start the run with MultiNA Control Software. See Figure 3 for the representative results for the Rluc-Y39×-Fluc reporter transcript.
Calculate the molar concentration of reporter mRNA as described below.
i. Take the ng/μL concentration by the result of the MultiNA. For example, since the concentration of Rluc-Y39×-Fluc with poly(A) tail in Figure 3 was 6.0 ng/μL, the stock solution should be 6.0 ng/μL × 10 (dilution factor from the stock) = 60 ng/μL.
ii. Estimate the poly(A) length by the result of MultiNA. For example, the poly(A) length of Rluc-Y39×-Fluc with a poly(A) tail in Figure 3 was ~100 nt.
iii. Calculate the molecular weight of reporter mRNA. Given that the non-poly(A)-tailed Rluc-Y39×-Fluc should be 2,787 nt, the poly(A)-tailed Rluc-Y39×-Fluc should be ~2,887 nt. Thus, the estimated molecular weight should be 2,887 × 320.5 = 0.93 × 106. For example, the molar concentration of poly(A)-tailed Rluc-Y39×-Fluc reporter in Figure 3 should be 60 [ng/μL]/(0.93 × 106) = 65 nM.
Dilute the reporter mRNA to 11 nM with RNase-free water.
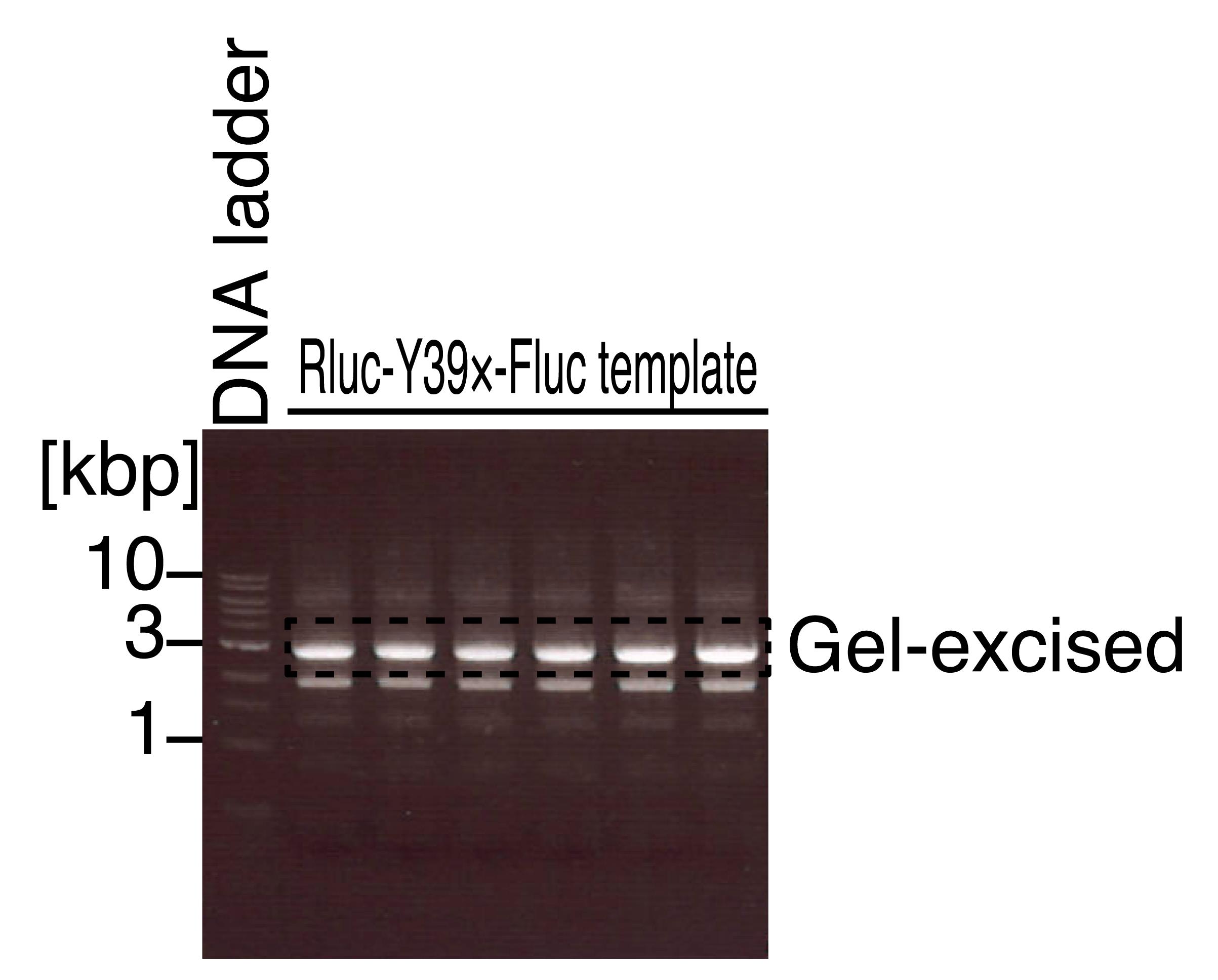
Figure 2. Agarose gel electrophoresis for the PCR-amplified DNA fragments for in vitro transcription templates. Representative gel image of the 1 kb DNA ladder and the PCR product for the in vitro transcription template of the Rluc-Y39×-Fluc reporter. The area indicated by the dashed line was gel excised. Note that this image was visualized by a UV transilluminator, which should be avoided for the actual gel-excision step.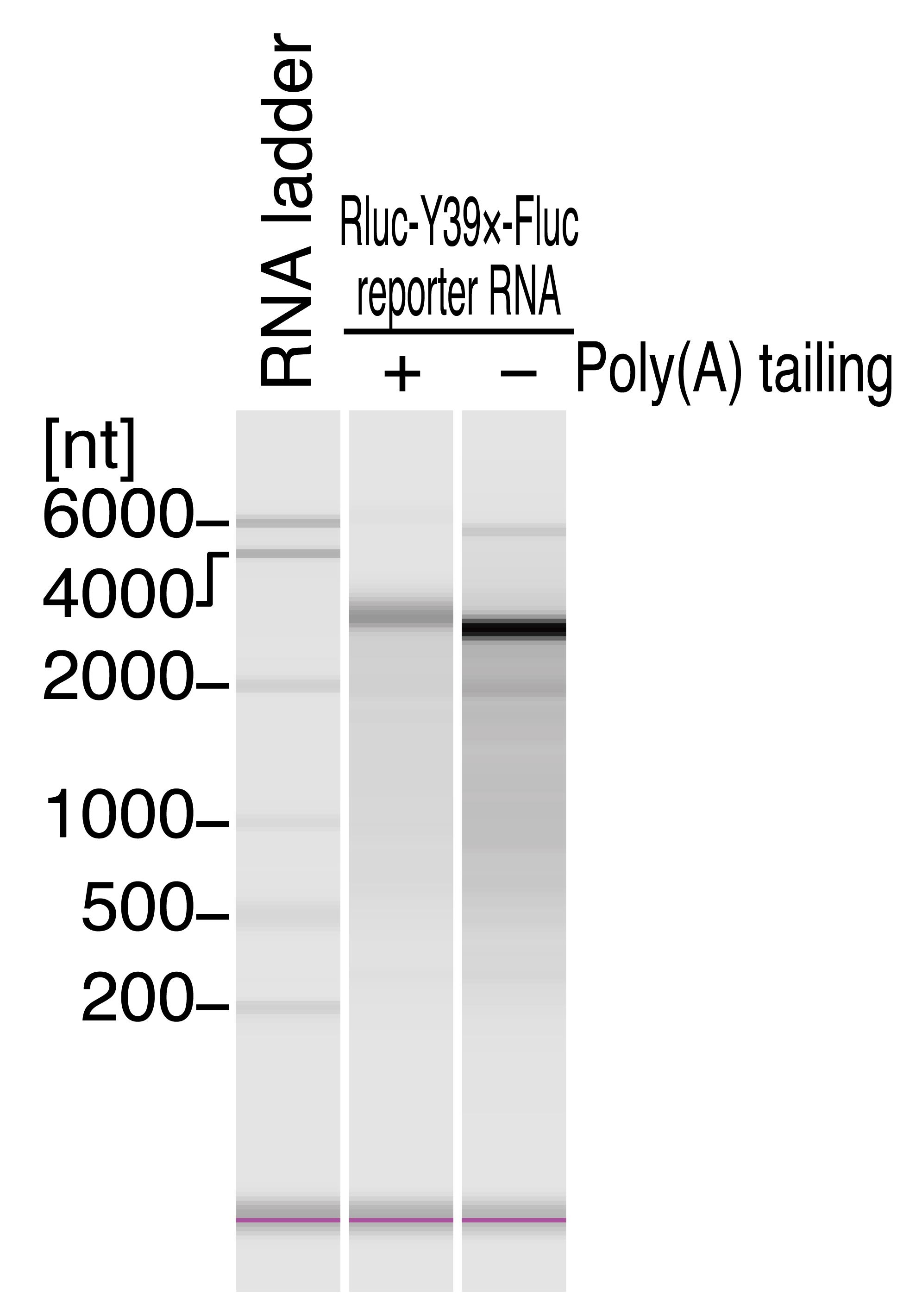
Figure 3. Quality check of the reporter transcript by fragment analyzer. Representative electropherograms of RNA 6000 ladder and Rluc-Y39×-Fluc reporter RNA with and without poly(A) tail. MultiNA Viewer software automatically assesses the size and concentration of RNA. The poly(A) tail length was estimated to be ~100 nt based on the length difference. Additionally, the RNA concentrations were determined as follows: Rluc-Y39×-Fluc with poly(A) tail, 6.0 ng/μL; Rluc-Y39×-Fluc without poly(A) tail, 23.3 ng/μL. We note that the concentration of individual mRNA measured by MultiNA may be lower than the total RNA concentration predicted by the DS-11 spectrophotometer.
Translation reaction
Prepare the in vitro translation reaction in a 1.5 mL DNA LoBind tube as described below (120 μL reaction).
Reagent Final concentration Amount Ribosome-depleted RRL n/a 60 μL 226 nM purified ribosome (from naïve or METTL18 KO cells) 22.6 nM 12 μL 11 nM reporter mRNA (Rluc-Y39×-Fluc or Rluc-Y0×-Fluc) 1.1 nM 12 μL Buffer KM (see Recipes) KCl: 75 mM
MgCl2: 0.75 mM
12 μL 200 μM amino acid mixture (see Recipes) 20 μM 12 μL 40 U/μL RNase inhibitor (TaKaRa) 0.8 U/μL 2.4 μL RNase-free water n/a 9.6 μL Note: RRL includes an energy regeneration system (phosphocreatine and phosphocreatine kinase), hemin, tRNA mixture, etc., which are generally required for in vitro translation. Thus, further supplementation of those materials is not necessary.
Start the reaction at 25 °C, take a 10 μL aliquot from the reaction every 5 min (for 50 min total, i.e., total 11 timepoints, including time 0 min) into 1.5 mL DNA LoBind tubes, immediately mix with 20 μL of 1× passive lysis buffer (see Recipes) to quench the reaction, and keep the mixture on ice until the luminescence measurement.
Measure the Rluc and Fluc luminescence by the Dual-Luciferase Reporter Assay System in Glomax.
Prepare Luciferase assay reagent II (LAR II) working solution as described below (for 44 + 1 samples).
Reagent Final concentration Amount Luciferase assay substrate (lyophilized product) n/a 1 vial Luciferase assay buffer II n/a 10 mL Notes:
i. Typically, the assay requires (number of samples +1) × 50 + 500 μL.
ii. The remaining LAR II can be stored at −80 °C for a month.
Prepare Stop & Glo working solution as described below.
Reagent Final concentration Amount 50× Stop & Glo substrate 1× 55 μL Stop & Glo buffer n/a 2,695 μL Note: Typically, the assay requires (number of samples +1) × 50 + 500 μL.
Wash both injectors of the GloMax Navigator System with Milli-Q water, 70% ethanol, Milli-Q water, and air, according to the manufacturer’s instructions.
Prime injectors with LAR II and Stop & Glo working solutions according to the manufacturer’s instructions.
Program the GloMax Navigator Software to perform a two-second premeasurement delay, followed by a 10-second measurement period for each assay, and to dispense 50 μL of LAR II and Stop & Glo working solutions per sample.
Transfer 20 μL of the reaction mixture (prepared in step 2 above) to each well of a white 96-well plate.
Place the white 96-well plate in the GloMax Navigator System and initiate the assay.
Export the luminescence data from the GloMax Navigator Software into a csv file. See the representative data of “GloMax_rawdata_1.csv” in Supplemental material.
Data analysis
Load the csv file into Excel software and analyze the data for the following steps. For the representative data analysis, see “Slope.xlsx” in Supplemental material. See Figure 4 for representative data analysis results.
Draw the scatter plots of Rluc and Fluc luminescence along the time.
Determine the linear part of the signal raise and calculate the slope (SlopeRluc and SlopeFluc).
Note: In this analysis, we used luminescence values at 25–45 min. The timepoint that provides a linear increase in the signal may depend on the experimental conditions.
Calculate SlopeFluc/SlopeRluc and then relative SlopeFluc/SlopeRluc (i.e., normalize the SlopeFluc/SlopeRluc of METTL18 KO ribosomes by that of naïve ribosomes).
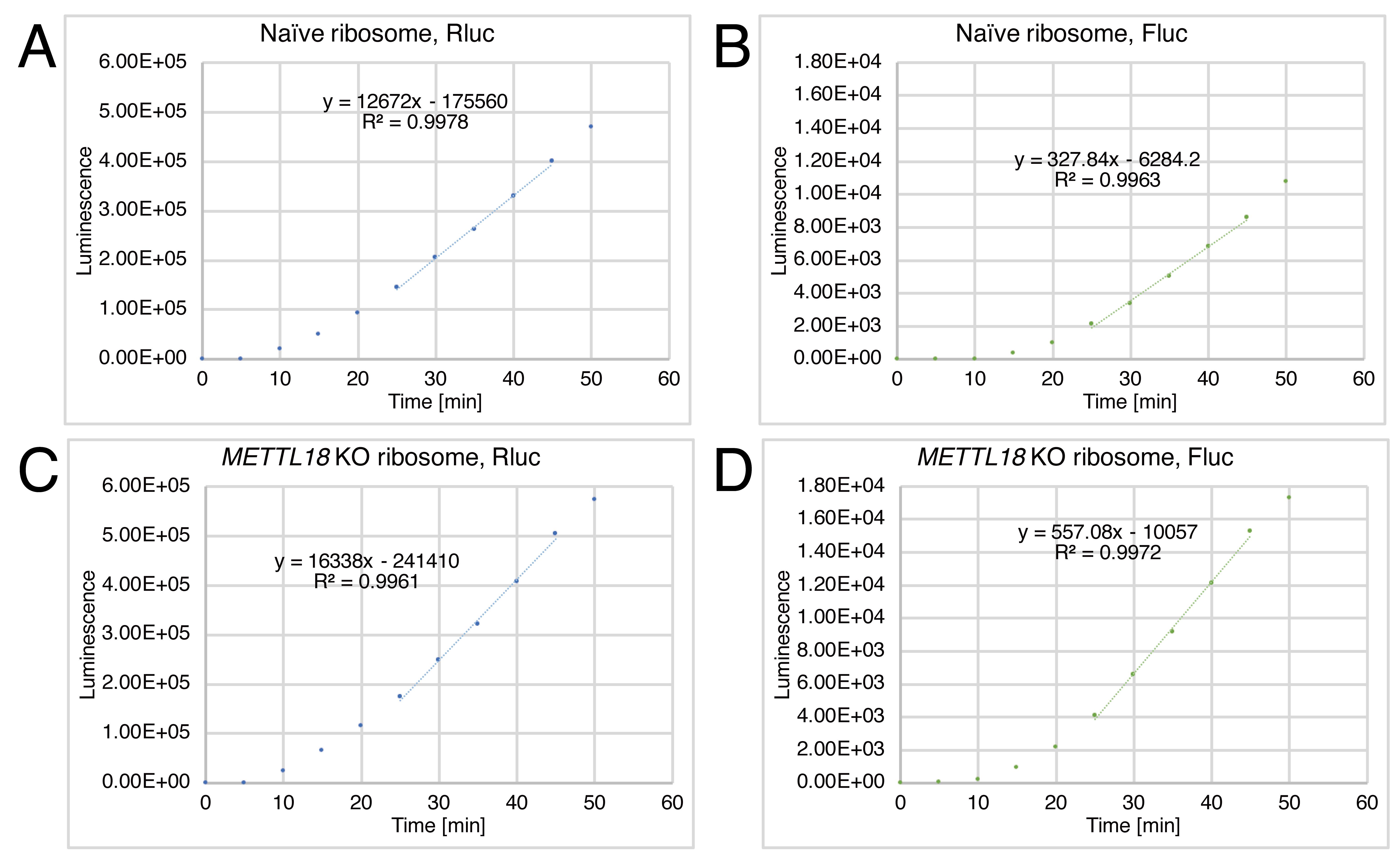
Figure 4. Representative data of the hybrid translation. Rluc and Fluc luminescence from the Rluc-Y39×-Fluc reporter over incubation time. Data for naïve ribosomes (A and B) and for METTL18 KO ribosomes (C and D) are shown. Based on the results, the SlopeFluc/SlopeRluc values for naïve ribosomes and METTL18 KO ribosomes were determined to be 0.0259 and 0.0341, respectively. Thus, the relative SlopeFluc/SlopeRluc for METTL18 KO ribosomes was 1.32, suggesting increased processivity of elongation along Y39×.
Notes
To test whether individual ribosome-depleted RRL and purified ribosomes have no translation activity on their own, small-scale reactions (10 μL) omitting each factor should be conducted prior to the experiments. See Figure 5 for the representative result, “GloMax_rawdata_2.csv” in Supplemental material for the raw data, and “Material_check.xlsx” in Supplemental material for the analysis.
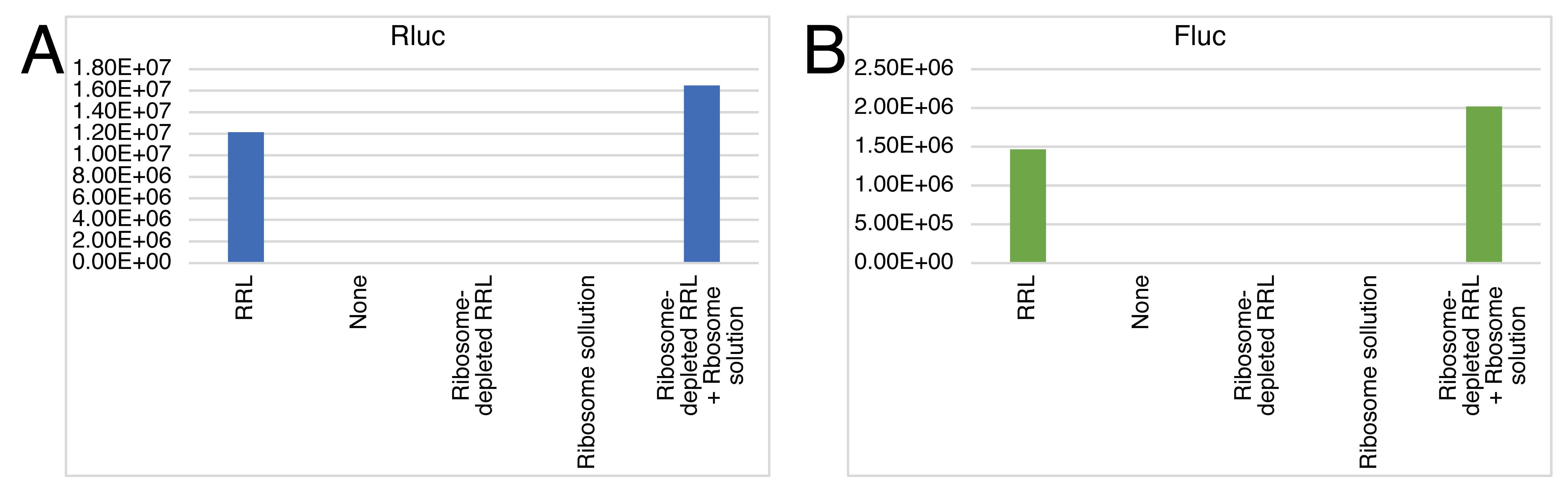
Figure 5. Control experiments for checking the quality of materials used in the hybrid translation. Rluc (A) and Fluc (B) luminescence from the Rluc-Y0×-Fluc reporter under the indicated conditions after 60 min of incubation. Data for naïve ribosomes are shown.
Recipes
DMEM supplemented with FBS (500 mL)
Reagent Final concentration Amount DMEM n/a 500 mL FBS 10% 50 mL Note: Keep at 4 °C.
1 M KOAc (5 mL)
Reagent Final concentration Amount KOAc 1 M 0.49 g RNase-free water n/a up to 5 mL Note: Keep at room temperature.
1 M MgOAc2 (1 mL)
Reagent Final concentration Amount MgOAc2·4H2O 1 M 0.214 g RNase-free water n/a up to 1 mL Note: Keep at room temperature.
1 M DTT (5 mL)
Reagent Final concentration Amount DTT 1 M 0.771 g RNase-free water n/a up to 5 mL Note: Store at -20 °C.
Buffer R (10 mL)
Reagent Final concentration Amount 1 M HEPES-KOH pH 7.5 10 mM 100 μL 1 M KOAc 10 mM 100 μL 1 M MgOAc2 1 mM 10 μL 1 M DTT 1 mM 10 μL RNase-free water n/a 9,780 μL Note: Prepare before use and keep on ice.
Sucrose cushion solution (10 mL for eight samples)
Reagent Final concentration Amount Sucrose 1 M 3.4 g (corresponding to 2.2 mL) 1 M HEPES-KOH pH 7.5 10 mM 100 μL 1 M KOAc 10 mM 100 μL 1 M MgOAc2 1 mM 10 μL 1 M DTT 1 mM 10 μL RNase-free water n/a 7,580 μL Note: Prepare before use and keep on ice.
Buffer R2 (5 mL)
Reagent Final concentration Amount 1 M HEPES-KOH pH 7.5 20 mM 100 μL 5 M NaCl 10 mM 10 μL 2 M KCl 25 mM 62.5 μL 1 M MgCl2 1.1 mM 5.5 μL 14.3 M 2-mercaptoethanol 7.7 mM 2.7 μL RNase-free water n/a 4819.3 μL Note: Prepare before use and keep on ice.
2× Laemmli sample buffer (1 mL)
Reagent Final concentration Amount 1 M Tris-HCl pH 6.8 125 mM 125 μL 100% Glycerol 20% 200 μL 10% SDS solution 4% 400 μL Bromophenol Blue 0.004% 0.04 mg RNase-free water n/a 175 μL 14.3 M 2-mercaptoethanol 10% 100 μL Notes:
Keep at room temperature (without 2-mercaptoethanol).
Add 2-mercaptoethanol before use.
10× SDS-PAGE running buffer (1 L)
Reagent Final concentration Amount Tris(hydroxymethyl)aminomethane 0.25 M 30.2 g Glycine 1.92 M 144 g Sodium lauryl sulfate 1% 10 g Milli-Q water n/a up to 1 L Note: Keep at room temperature.
1× SDS-PAGE running buffer (1 L)
Reagent Final concentration Amount 10× Running buffer 1× 100 mL Milli-Q water n/a 900 mL Note: Keep at room temperature.
Gel fixation buffer (100 mL)
Reagent Final concentration Amount Methanol 50% 50 mL Acetic acid 7% 7 mL Milli-Q water n/a 43 mL Note: Keep at room temperature.
50× TAE (1 L)
Reagent Final concentration Amount Tris(hydroxymethyl)aminomethane 2 M 242 g Acetic acid 1 M 57.1 mL 0.5 M EDTA (pH 8.0) 50 mM 100 mL Milli-Q water n/a up to 1 L 1% agarose gel (100 mL)
Reagent Final concentration Amount Agarose 1% 1 g 50× TAE 1× 2 mL Milli-Q water n/a 97 mL Note: Dissolve agarose by heating with a microwave oven. Pour the reagent into the gasket and cool it until use.
70% ethanol (50 mL)
Reagent Final concentration Amount Ethanol 70% 35 mL RNase-free water n/a 15 mL Buffer KM (500 μL)
Reagent Final concentration Amount 2 M KCl 750 mM 187 μL 1 M MgCl2 7.5 mM 3.7 μL RNase-free water n/a 309.3 μL Note: Prepare before use and keep on ice.
200 μM amino acid mixture (500 μL)
Reagent Final concentration Amount 1 mM amino acid mixtures 200 μM 100 μL RNase-free water n/a 400 μL Note: Store at -80 °C.
1× passive lysis buffer (5 mL)
Reagent Final concentration Amount 5× passive lysis buffer 1× 1 mL RNase-free water n/a 4 mL Note: Prepare before use.
Acknowledgments
We are grateful to all the members of the Iwasaki laboratory for technical help. This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (a Grant-in-Aid for Transformative Research Areas [B] “Parametric Translation”, JP20H05784 to S.I.); the Japan Society for the Promotion of Science (JSPS) (a Grants-in-Aid for Young Scientists [A], JP17H04998 to S.I.; a Grant-in-Aid for Scientific Research [C], JP21K06026 to E.M.S.; a Grant-in-Aid for Research Activity start-up, JP22K20765 to H.T.); the Japan Agency for Medical Research and Development (AMED) (AMED-CREST, JP22gm1410001 to S.I.); RIKEN (Pioneering Projects “Biology of Intracellular Environments” to S.I.; Collaboration Seed Fund to E.M.S. ; Incentive Research Project to E.M.S. and H.T.). This protocol was derived from an original paper (Matsuura-Suzuki et al., 2022) and the manufacturer’s protocols for the Qubit RNA BR Assay kit (Thermo Fisher Scientific), NucleoSpin Gel and PCR Clean-up (MACHEREY-NAGEL), T7-Scribe Standard RNA IVT kit (CELLSCRIPT), ScriptCap m7G Capping system (CELLSCRIPT), ScriptCap 2′-O-Methyltransferase Kit (CELLSCRIPT), A-Plus Poly(A) Polymerase Tailing Kit (CELLSCRIPT), Dual-Luciferase Reporter Assay System (Promega), and GloMax Navigator System with Dual Injectors (Promega).
Competing interests
The authors declare no competing interests.
Ethical considerations
No human or animal subjects were included in this study.
References
- Abe, T., Nagai, R., Shimazaki, S., Kondo, S., Nishimura, S., Sakaguchi, Y., Suzuki, T., Imataka, H., Tomita, K. and Takeuchi-Tomita, N. (2020). In vitro yeast reconstituted translation system reveals function of eIF5A for synthesis of long polypeptide. J Biochem 167(5): 451-462.
- Alkalaeva, E. Z., Pisarev, A. V., Frolova, L. Y., Kisselev, L. L. and Pestova, T. V. (2006). In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125(6): 1125-1136.
- Bergamini, G., Preiss, T. and Hentze, M. W. (2000). Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6(12): 1781-1790.
- Brödel, A. K., Sonnabend, A. and Kubick, S. (2014). Cell-free protein expression based on extracts from CHO cells. Biotechnol Bioeng 111(1): 25-36.
- Dieterich, D. C., Hodas, J. J., Gouzer, G., Shadrin, I. Y., Ngo, J. T., Triller, A., Tirrell, D. A. and Schuman, E. M. (2010). In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 13(7): 897-905.
- Emmott, E., Jovanovic, M. and Slavov, N. (2019). Ribosome stoichiometry: From form to function. Trends Biochem Sci 44(2): 95-109.
- Erales, J., Marchand, V., Panthu, B., Gillot, S., Belin, S., Ghayad, S. E., Garcia, M., Laforêts, F., Marcel, V., Baudin-Baillieu, A., et al. (2017). Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc Natl Acad Sci U S A 114(49): 12934-12939.
- Ferretti, M. B. and Karbstein, K. (2019). Does functional specialization of ribosomes really exist? RNA 25(5): 521-538.
- Fritz, S. E., Haque, N. and Hogg, J. R. (2018). Highly efficient in vitro translation of authentic affinity-purified messenger ribonucleoprotein complexes. RNA 24(7): 982-989.
- Genuth, N. R. and Barna, M. (2018a). Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat Rev Genet 19(7): 431-452.
- Genuth, N. R. and Barna, M. (2018b). The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol Cell 71(3): 364-374.
- Gregorio, N. E., Levine, M. Z. and Oza, J. P. (2019). A User’s Guide to Cell-Free Protein Synthesis. Methods Protoc 2(1).
- Guo, H. (2018). Specialized ribosomes and the control of translation. Biochem Soc Trans 46(4): 855-869.
- Hunt, T. and Jackson, R. J. (1974). The rabbit reticulocyte lysate as a system for studying mRNA. Hamatol Bluttransfus 14: 300-307.
- Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. and Weissman, J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324(5924): 218-223.
- Iwasaki, S. and Ingolia, N. T. (2017). The growing toolbox for protein synthesis studies. Trends Biochem Sci 42(8): 612-624.
- Jackson, R. J. and Hunt, T. (1983). Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol 96: 50-74.
- Kerr, I. M., Cohen, N. and Work, T. S. (1966). Factors controlling amino acid incorporation by ribosomes from krebs II mouse ascites-tumour cells. Biochem J 98(3): 826-835.
- Kisly, I., Remme, J. and Tamm, T. (2018). Ribosomal protein eL24, involved in two intersubunit bridges, stimulates translation initiation and elongation. Nucleic Acids Res 47(1): 406-420.
- Kisly, I., Kattel, C., Remme, J. and Tamm, T. (2021). Luciferase-based reporter system for in vitro evaluation of elongation rate and processivity of ribosomes. Nucleic Acids Res 49(10): e59.
- Machida, K., Shigeta, T., Yamamoto, Y., Ito, T., Svitkin, Y., Sonenberg, N. and Imataka, H. (2018). Dynamic interaction of poly(A)-binding protein with the ribosome. Sci Rep 8(1): 17435.
- Małecki, J. M., Odonohue, M.-F., Kim, Y., Jakobsson, M. E., Gessa, L., Pinto, R., Wu, J., Davydova, E., Moen, A., Olsen, J. V., et al. (2021). Human METTL18 is a histidine-specific methyltransferase that targets RPL3 and affects ribosome biogenesis and function. Nucleic Acids Res 49(6): 3185-3203.
- Mathews, M. B. and Korner, A. (1970). Mammalian Cell-Free Protein Synthesis Directed by Viral Ribonucleic Acid. Eur J Biochem 17(2): 328-338.
- Matsuura-Suzuki, E., Shimazu, T., Takahashi, M., Kotoshiba, K., Suzuki, T., Kashiwagi, K., Sohtome, Y., Akakabe, M., Sodeoka, M., Dohmae, N., et al. (2022). METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance. Elife 11: e72780.
- Mikami, S., Masutani, M., Sonenberg, N., Yokoyama, S. and Imataka, H. (2006). An efficient mammalian cell-free translation system supplemented with translation factors. Protein Expr Purif 46(2): 348-357.
- Molla, A., Paul, A. V. and Wimmer, E. (1991). Cell-free, de novo synthesis of poliovirus. Science 254(5038): 1647-1651.
- Morisaki, T., Lyon, K., DeLuca, K. F., DeLuca, J. G., English, B. P., Zhang, Z., Lavis, L. D., Grimm, J. B., Viswanathan, S., Looger, L. L., et al. (2016). Real-time quantification of single RNA translation dynamics in living cells. Science 352(6292): 1425-1429.
- Panthu, B., Décimo, D., Balvay, L. and Ohlmann, T. (2015). In vitro translation in a hybrid cell free lysate with exogenous cellular ribosomes. Biochem J 467(3): 387-398.
- Pelham, H. R. B. and Jackson, R. J. (1976). An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem 67(1): 247-256.
- Penzo, M., Rocchi, L., Brugiere, S., Carnicelli, D., Onofrillo, C., Couté, Y., Brigotti, M. and Montanaro, L. (2015). Human ribosomes from cells with reduced dyskerin levels are intrinsically altered in translation. FASEB J 29(8): 3472-3482.
- Penzo, M., Carnicelli, D., Montanaro, L. and Brigotti, M. (2016). A reconstituted cell-free assay for the evaluation of the intrinsic activity of purified human ribosomes. Nat Protoc 11(7): 1309-1325.
- Pestova, T. V., Borukhov, S. I. and Hellen, C. U. (1998). Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394(6696): 854-859.
- Pestova, T. V. and Hellen, C. U. T. (2003). Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev 17(2): 181-186.
- Pisarev, A. V., Hellen, C. U. T. and Pestova, T. V. (2007). Recycling of eukaryotic posttermination ribosomal complexes. Cell 131(2): 286-299.
- Rakotondrafara, A. M. and Hentze, M. W. (2011). An efficient factor-depleted mammalian in vitro translation system. Nat Protoc 6(5): 563-571.
- Schwanhäusser, B., Gossen, M., Dittmar, G. and Selbach, M. (2009). Global analysis of cellular protein translation by pulsed SILAC. Proteomics 9(1): 205-209.
- Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K. and Ueda, T. (2001). Cell-free translation reconstituted with purified components. Nat Biotechnol 19(8): 751-755.
- Simsek, D. and Barna, M. (2017). An emerging role for the ribosome as a nexus for post-translational modifications. Curr Opin Cell Biol 45: 92-101.
- Svitkin, Y. V. and Agol, V. I. (1978). Complete translation of encephalomyocarditis virus RNA and faithful cleavage of virus-specific proteins in a cell-free system from Krebs-2 cells. FEBS Lett 87(1): 7-11.
- Svitkin, Y. V. and Sonenberg, N. (2004). An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol Biol 257: 155-170.
- Svitkin, Y. V. and Sonenberg, N. (2007). A highly efficient and robust in vitro translation system for expression of picornavirus and hepatitis C virus RNA genomes. Methods Enzymol 429: 53-82.
- Taoka, M., Nobe, Y., Yamaki, Y., Sato, K., Ishikawa, H., Izumikawa, K., Yamauchi, Y., Hirota, K., Nakayama, H., Takahashi, N., et al. (2018). Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res 46(18): 9289-9298.
- Thoma, C., Ostareck-Lederer, A. and Hentze, M. W. (2004). A poly(A) tail-responsive in vitro system for cap- or IRES-driven translation from HeLa cells. Methods Mol Biol 257: 171-180.
- Trainor, B. M., Pestov, D. G. and Shcherbik, N. (2021). Development, validation, and application of the ribosome separation and reconstitution system for protein translation in vitro. RNA 27(12): 1602-1616.
- Wang, C., Han, B., Zhou, R. and Zhuang, X. (2016). Real-time imaging of translation on single mRNA transcripts in live cells. Cell 165(4): 990-1001.
- Witherell, G. (2001). In vitro translation using HeLa extract. Curr Protoc Cell Biol Chapter 11: Unit 11.8. doi: 10.1002/0471143030.cb1108s06.
- Wu, B., Eliscovich, C., Yoon, Y. J. and Singer, R. H. (2016). Translation dynamics of single mRNAs in live cells and neurons.Science 352(6292): 1430-1435.
- Yan, X., Hoek, T. A., Vale, R. D. and Tanenbaum, M. E. (2016). Dynamics of translation of single mRNA molecules in vivo. Cell 165(4): 976-989.
- Yokoyama, T., Machida, K., Iwasaki, W., Shigeta, T., Nishimoto, M., Takahashi, M., Sakamoto, A., Yonemochi, M., Harada, Y., Shigematsu, H., et al. (2019). HCV IRES captures an actively translating 80S ribosome. Mol Cell 74(6): 1205-1214.e8.
Supplementary information
The following supporting information can be downloaded here:
- GloMax_rawdata_1.csv
- GloMax_rawdata_2.csv
- Slope.xlsx
- Material_check.xlsx
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Matsuura-Suzuki, E., Toh, H. and Iwasaki, S. (2023). Human-rabbit Hybrid Translation System to Explore the Function of Modified Ribosomes. Bio-protocol 13(13): e4714. DOI: 10.21769/BioProtoc.4714.
- Matsuura-Suzuki, E., Shimazu, T., Takahashi, M., Kotoshiba, K., Suzuki, T., Kashiwagi, K., Sohtome, Y., Akakabe, M., Sodeoka, M., Dohmae, N., et al. (2022). METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance. Elife 11: e72780.
分类
生物化学 > RNA > mRNA翻译
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












