- EN - English
- CN - 中文
easyPACId, a Simple Method for Induced Production, Isolation, Identification, and Testing of Natural Products from Proteobacteria
easyPACId,一种诱导生产、分离、鉴定和测试变形杆菌天然产物的简单方法
(*contributed equally to this work) 发布: 2023年07月05日第13卷第13期 DOI: 10.21769/BioProtoc.4709 浏览次数: 1620
评审: Dennis J NürnbergSelcuk HazirAnonymous reviewer(s)
Abstract
The easyPACId (easy Promoter Activation and Compound Identification) approach is focused on the targeted activation of natural product biosynthetic gene clusters (BGCs) encoding non-ribosomal peptide synthetases (NRPS), polyketide synthases (PKS), NRPS-PKS hybrids, or other BGC classes. It was applied to entomopathogenic bacteria of the genera Xenorhabdus and Photorhabdus by exchanging the natural promoter of desired BGCs against the L-arabinose inducible PBAD promoter in ∆hfq mutants of the respective strains. The crude (culture) extracts of the cultivated easyPACId mutants are enriched with the single compound or compound class and can be tested directly against various target organisms without further purification of the produced natural products. Furthermore, isolation and identification of compounds from these mutants is simplified due to the reduced background in the ∆hfq strains. The approach avoids problems often encountered in heterologous expression hosts, chemical synthesis, or tedious extraction of desired compounds from wild-type crude extracts. This protocol describes easyPACId for Xenorhabdus and Photorhabdus, but it was also successfully adapted to Pseudomonas entomophila and might be suitable for other proteobacteria that carry hfq.
Keywords: easyPACId (easyPACId)Background
Entomopathogenic bacteria of the genera Xenorhabdus and Photorhabdus live in symbiosis with soil dwelling nematodes of the genera Steinernema or Heterorhabditis, respectively (Goodrich-Blair and Clarke, 2007; Chaston et al., 2011). They are able to propagate without their nematode host and can be easily cultivated in the laboratory using standard media. Their genome harbors a rich diversity for natural products (NPs) with potential activities as antibiotics, antifungals, and cytotoxic or signaling functions. These NPs are often produced by non-ribosomal peptide synthetases (NRPS), polyketide synthases (PKS), and mixed NRPS-PKS hybrids, encoded by biosynthetic gene clusters (BGCs) (H. B. Bode, 2009; Shi and Bode, 2018; Shi et al., 2022).
The access to this promising source of bioactive compounds is possible via different approaches: either by optimizing the culture conditions for enhanced NP production, by heterologous expression or promoter activation of the BGC of interest, or chemical synthesis of the NP derived thereof (H. B. Bode et al., 2002; Biggins et al., 2011; Biggins et al., 2014; E. Bode et al., 2017). All these methods have their limitations: chemical synthesis might be impossible, difficult, or too time consuming, requiring profound knowledge of the NP structure as a starting point. Heterologous production of the desired NP is dependent on building blocks that might only be present in the natural host, on the size of the BGC to be cloned into the heterologous host, on its toxicity, or on special transport systems or modifying enzymes (Brachmann et al., 2012; Nollmann et al., 2015). Isolation of natural products from crude extracts from wildtype (WT) bacteria might be hindered by low production titers, regulatory elements that silence the expression of BGCs, and/or sophisticated culture conditions that have to be established. A promoter activation of desired gene clusters in WT strains might result in NP overproduction that facilitates the isolation of the compound for bioactivity testing and structure elucidation, but still requires NP isolation from crude extracts. The easyPACId (easy Promoter Activation and Compound Identification) approach circumvents all these limitations.
easyPACId describes a simple and fast enrichment and preparation of selected NPs for bioactivity screening just from the crude extract of a bacterial culture of an induced ∆hfq_pCEP-gene_xyz easyPACId mutant. It avoids laborious steps often encountered during NP isolation from WT crude extracts and is not hindered by limitations concerning heterologous expression or chemical synthesis. The targeted activation via the PBAD promoter (Guzman et al., 1995) in a clean ∆hfq background serves as an excellent tool to enable production of only desired NPs or NP classes derived from single BGCs (Tobias et al., 2017; Valverde, 2017; E. Bode et al., 2019). The RNA chaperone Hfq is widespread in proteobacteria. It has a pleiotropic effect on gene expression by mediating the interaction of small non-coding RNAs (sRNA) and mRNAs, and thereby can activate or suppress translation (Vogel and Luisi, 2011). For NP-producing bacteria like Xenorhabdus, Photorhabdus, or Pseudomonas, a ∆hfq mutant has lost the ability to produce all or most NPs or shows a severely reduced NP amount. Consequently, the chromatogram of crude extracts from a ∆hfq mutant analyzed via HPLC/MS shows no NP-derived signals anymore (clean background) compared to HPLC/MS chromatograms of the corresponding WT where all NPs are produced, and their corresponding signals show a typical pattern for the strain (Figure 1a).
Briefly, the experiment starts via detection of potential BGCs encoding NRPSs, PKS, hybrids thereof, or other BGC classes, via antiSMASH analysis (or based on other bioinformatic tools) (Medema et al., 2011) in the genome of selected NP producers, here exemplified for the genera Photorhabdus and Xenorhabdus. By deletion of the gene hfq, encoding the so-called RNA-chaperone Hfq, a mutant is generated, which is deficient in NP production (Tobias et al., 2017). By exchange of the natural promoter against the L-arabinose inducible promoter PBAD (Guzman et al., 1995), encoded on the conjugatable vector pCEP via homologous integration (E. Bode et al., 2015), the selective activation of the targeted BGC in a ∆hfq background is now possible (Figure 1). Cell-free culture supernatants or even whole cultures from respective ∆hfq_pCEP-gene_xyz can be freeze dried and dissolved in water for subsequent testing on various target organisms (E. Bode et al., 2015 and 2019; Gulsen et al., 2022). Often, the selected compounds are overproduced and can be easily isolated for further investigations like structure elucidation.
This simple and beneficial method of gaining potentially bioactive compounds can be used to activate natural product biosynthesis gene clusters, not only in Xenorhabdus and Photorhabdus bacteria. It has been applied already to Pseudomonas and is likely to work in any other proteobacteria that encode Hfq, like Serratia or Vibrio (Thoma and Schobert, 2009; Hmelo et al., 2015). Besides Hfq itself, other regulative components of the hfq-sRNA regulon like ArcZ (Neubacher et al., 2020) or even other global regulators might also be useful to generate NP non-producing phenotypes in the described or other Gram-negative bacteria.
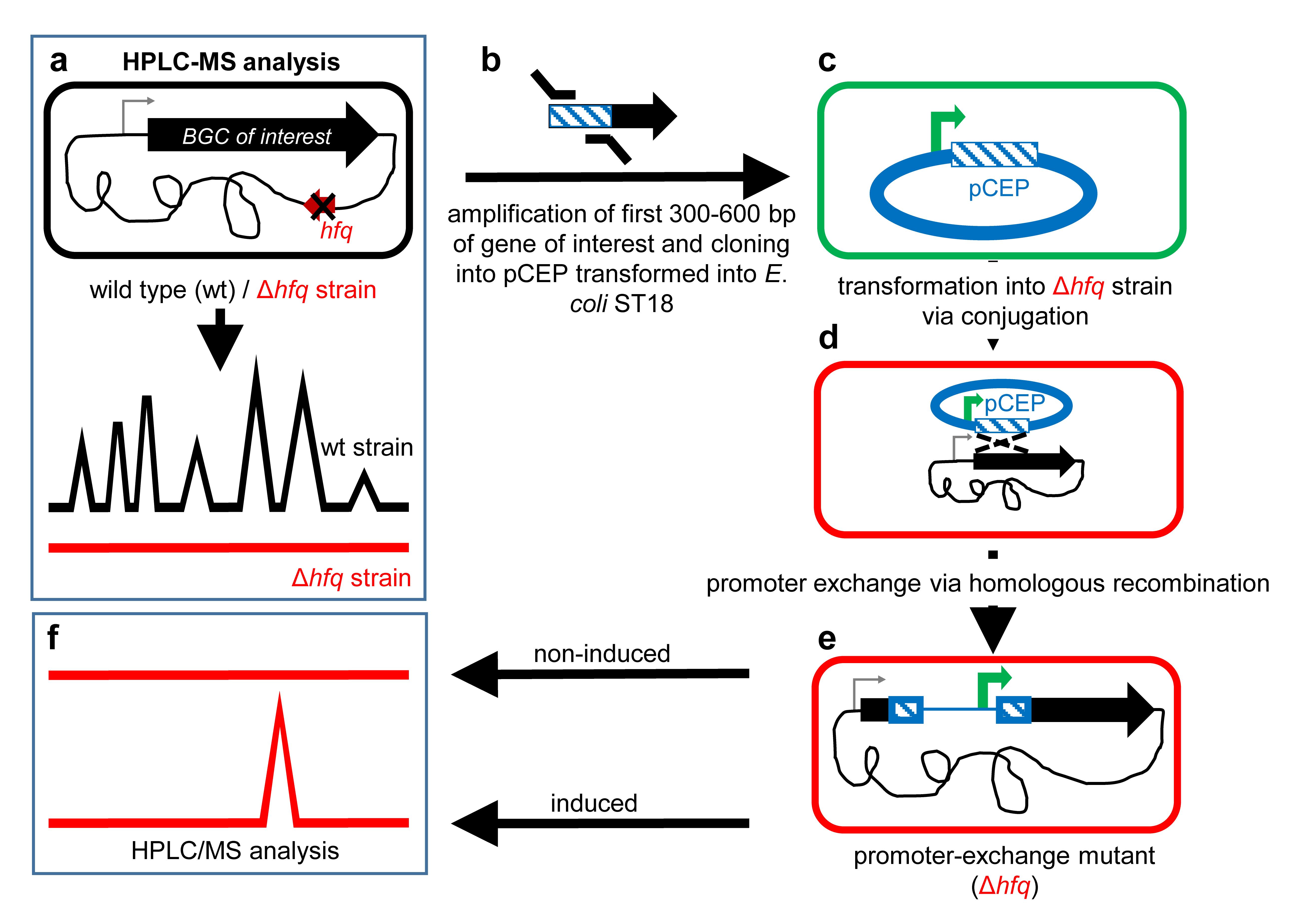
Figure 1. Schematic overview of the easyPACId workflow
Materials and reagents
∆hfq strain from Xenorhabdus spp. (E. Bode et al., 2019)
Sterile tips for micropipettes [Starlab, catalog numbers: S1110-3700-c (10–20 μL), S1111-1706-c (200 μL), S1111-6000-c (1,000 μL)]
Sterile reaction tubes [Sarstedt, catalog numbers: 72.706 (1.5 mL), 72.695.500 (2.0 mL)]
Black reaction tubes (Carl Roth, catalog number: AA80.1)
PCR tubes (Starlab, catalog number: 1402-3700)
50 mL tubes (Sarstedt, catalog number: 62.547.254)
Petri dishes [Sarstedt, catalog numbers: 82.1473.001 (92 mm × 16 mm), 82.1184.500 (150 mm × 20 mm)]
Electroporation cuvettes (Bio-Rad, catalog number: 1652089)
Single-use syringe Omnifix LuerLock Solo (Braun, catalog number: 4617207V)
Filtropur S 0.2 (Sarstedt, catalog number: 83.1826.001)
Steritop 45 mm neck size Millipore Express Plus 0.22 μm PES (Millipore, catalog number: S2GPT05RE)
Tryptone (BD, catalog number: 211699)
Yeast extract (BD, catalog number: 288620)
Sodium chloride (NaCl) (Carl Roth, catalog number: 3957.2)
Bacto-agar (BD, catalog number: 214030)
LE agarose (Biozym, catalog number: 840004)
L-Arabinose (Carl Roth, catalog number: 5118.5)
Sodium hydroxide (NaOH) (Carl Roth, catalog number: 6771.1)
Kanamycin monosulfate (km) (Sigma, catalog number: K4000)
5-Aminolevulinic acid hydrochloride (5-ALA) (Sigma, catalog number: 8339)
E. coli ST18 competent cells (Thoma and Schobert, 2009) (DSMZ, Braunschweig, Germany)
Conjugatable cloning vector pCEP-kmR with R6K ori, PBAD promoter, selection marker kanamycin (kmR) (E. Bode et al., 2015 and 2019)
Phusion High-Fidelity DNA Polymerase (New England Biolabs, catalog number: M0530L)
dNTP-Set (Carl Roth, catalog number: K039.2)
PstI/PstI HF (New England Biolabs, catalog numbers: R0140L/R3140L)
NdeI (New England Biolabs, catalog number: R0111L)
Monarch Genomic DNA Purification kit (New England Biolabs, catalog number: T3010L)
100 bp DNA ladder (New England Biolabs, catalog number: N3231L); 1 kb DNA ladder (New England Biolabs, catalog number: N3232L)
Midori Green Advance (Biozym, catalog number: 617004)
QIAprep Spin Miniprep kit (Qiagen, catalog number: 27106)
Wizard® SV Gel and PCR Clean-Up System (Promega, catalog number: A9285)
NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, catalog number: E2621L)
Methanol optima LC/MS grade (Fisher Scientific, catalog number: A456-212)
Ethanol (Carl Roth, catalog number: 9065.4)
Glycerol (Carl Roth, catalog number: 3783.2)
Acetic acid (Carl Roth, catalog number: 3738.5)
Trizma-Base (Sigma, catalog number: T6066)
Na2-EDTA (Carl Roth, catalog number: 8043.2)
Luria-Bertani medium (LB) (see Recipes)
5-Aminolevulinic acid hydrochloride (5-ALA) (50 mg/mL) (see Recipes)
L-Arabinose (20% w/v) (see Recipes)
Kanamycin (50 mg/mL) (see Recipes)
TAE-buffer, pH 8.0 (see Recipes)
0.5 M EDTA pH 8.0 (see Recipes)
Equipment
Super Duty Erlenmeyer flasks, 50 mL (Omnilab, catalog number: 5425046)
500 mL Erlenmeyer flasks (Omnilab, catalog number: 5010476)
Cellulose plug F 17.0–18.5 mm (VWR International, catalog number: 391-0161)
Caps for flasks with neck Ø38 mm (Carl Roth, catalog number: K396.1)
Sterile glass pipettes
Gel chambers for agarose gel electrophoresis
Inoculation loop, Platin-Iridium (neoLab, catalog number: 1-2112)
Drigalski spatula (Carl Roth, catalog number: T724.2)
Sterile toothpicks
Conical centrifuge tube 50 mL (Sarstedt, catalog number: 62.547.254)
ChemiDoc MP imaging systems with Lab Touch Software (Bio-Rad, catalog number: 12003154) and Blot/UV/Stain-Free tray (Bio-Rad, catalog number: 12003028)
Transilluminator (Intas Science Imaging Instruments)
Micropipettes (Gilson, Pipetman G P2G, catalog number: F144054M; Pipetman G P10G, catalog number: F144055M; Pipetman G P20G, catalog number: F144056M; Pipetman G P200G, catalog number: F144058M; Pipetman G P1000G, M, catalog number: F144059M)
Centrifuge Pico 17 microcentrifuge (Fisher Scientific, catalog number: 75002410)
Centrifuge Eppendorf 5810 R (V8.8), rotor S-4-104 with adapter (Eppendorf, catalog numbers: 5825734009 and 5825733002)
ProFlexTM PCR system, 96-well (Applied BiosystemsTM, catalog number: 4484075)
Eppendorf ThermoMixer C (Eppendorf, catalog number: 5382000015)
Infors HT-Multitron Standard shaker
Gene Pulser Xcell (Bio-Rad)
NanoDrop One WiFi (Life Technologies, catalog number: ND-ONE-W)
UV-vis-spectrophotometer Biochrom Libra S60 (Omnilab, catalog number: 80-7000-12)
Lyovapor L-300 (Büchi)
HPLC vials X1000 Microflaschen ND9 (Fisher Scientific, catalog number: 11707597)
Lid (for HPLC vials) X1000 (Fisher Scientific, catalog number: 11787567)
HPLC/MS Thermo Scientific UltiMate 3000 coupled to an amaZon speed ion trap (Bruker)
Vortex Genie 2 (Fisher Scientific, catalog number: 15547335)
Software
Geneious Prime® 2022.1.1 or earlier versions (https://www.geneious.com)
DataAnalysis (Bruker Daltonics GmbH)
antiSMASH (https://antismash.secondarymetabolites.org/#!/start) (Medema et al., 2011)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Bode, E., Assmann, D., Happel, P., Meyer, E., Münch, K., Rössel, N. and Bode, H. B. (2023). easyPACId, a Simple Method for Induced Production, Isolation, Identification, and Testing of Natural Products from Proteobacteria. Bio-protocol 13(13): e4709. DOI: 10.21769/BioProtoc.4709.
分类
微生物学 > 微生物生理学
生物工程 > 合成生物学
分子生物学 > DNA
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link