- EN - English
- CN - 中文
Qualitative and Quantitative Methods to Measure Antibacterial Activity Resulting from Bacterial Competition
测定细菌竞争产生的抗菌活性的定性和定量方法
发布: 2023年07月05日第13卷第13期 DOI: 10.21769/BioProtoc.4706 浏览次数: 3517
评审: Cristina Alvarez MartinezAksiniya AsenovaAnonymous reviewer(s)
Abstract
In the environment, bacteria compete for niche occupancy and resources; they have, therefore, evolved a broad variety of antibacterial weapons to destroy competitors. Current laboratory techniques to evaluate antibacterial activity are usually labor intensive, low throughput, costly, and time consuming. Typical assays rely on the outgrowth of colonies of prey cells on selective solid media after competition. Here, we present fast, inexpensive, and complementary optimized protocols to qualitatively and quantitively measure antibacterial activity. The first method is based on the degradation of a cell-impermeable chromogenic substrate of the β-galactosidase, a cytoplasmic enzyme released during lysis of the attacked reporter strain. The second method relies on the lag time required for the attacked cells to reach a defined optical density after the competition, which is directly dependent on the initial number of surviving cells.
Key features
• First method utilizes the release of β-galactosidase as a proxy for bacterial lysis.
• Second method is based on the growth timing of surviving cells.
• Combination of two methods discriminates between cell death and lysis, cell death without lysis, or survival to quasi-lysis.
• Methods optimized to various bacterial species such as Escherichia coli, Pseudomonas aeruginosa, and Myxococcus xanthus.
Graphical overview
Background
In the environment, bacteria do not live alone but rather build complex microbial communities in which several species coexist (Luo et al., 2022). These microbial communities, while dynamic, represent ecosystems that are usually stable and resilient by resisting changing conditions and disturbances. To ensure this stability, bacteria establish relationships with other bacterial species and microbes for mutual benefits. The most studied cooperation mechanisms are quorum sensing (i.e., a communication mechanism based on release and sensing of signaling molecules allowing the synchronized expression of specific genes), exchange of metabolic molecules, and division of labor (subpopulations that perform different tasks simultaneously) (West et al., 2006). Conversely, antagonist interactions, collectively known as competition, correspond to mechanisms evolved by microorganisms to destroy rivals—or prevent their growth—in order to ensure a privileged access to the niche or to limited resources. Competition can lead to the increase of one species at the expense of one or several others, and usually promotes formation of spatial segregation patterns inside the community. Competition comprises indirect (exploitative) or direct (interference) mechanisms, in which one species consumes the resources or eliminate rivals, respectively (Ghoul and Mitri, 2016). An attacker cell competing for nutrients or space is referred as a competitor. By opposition, an attacker using competition to directly eliminate and feed from a target cell is referred as a predator. At the molecular level, many mechanisms involved in direct competition have been revealed and described in detail (Ghoul and Mitri, 2016; Stubbendieck and Straight, 2016; Granato et al., 2019). All these mechanisms rely on common themes but need distinct specific machineries. They all require the delivery of a potent toxin that is usually co-produced with a cognate immunity protein conferring protection to the attacker cell (Chassaing and Cascales, 2018). Toxin-mediated competition mechanisms are categorized as contact-independent or contact-dependent. The best-known example of contact-independent mechanisms is represented by bacteriocins, such as colicins. These are peptide or protein antibiotics that are released by a quasi-lysis mechanism and bind to and penetrate into target cells by hijacking essential components of the cell envelope (Cascales et al., 2007; Kleanthous, 2010). By contrast, contact-dependent mechanisms rely on specific secretion machineries required for cell surface exposure or the delivery of toxic effectors. These are either directly injected inside the target cell or use a penetration mechanism similar to that of bacteriocins. Several dedicated secretion systems, such as Type IV, Type V, Type VI, and Type VII secretion systems and Type IV and Tad pili, have been demonstrated to play key roles in bacterial competition (Ruhe et al., 2013; Willett et al., 2015; Cao et al., 2016; Chassaing and Cascales, 2018; Granato et al., 2019; Sgro et al., 2019; Seef et al., 2021; Tassinari et al., 2022; Thiery et al., 2022).
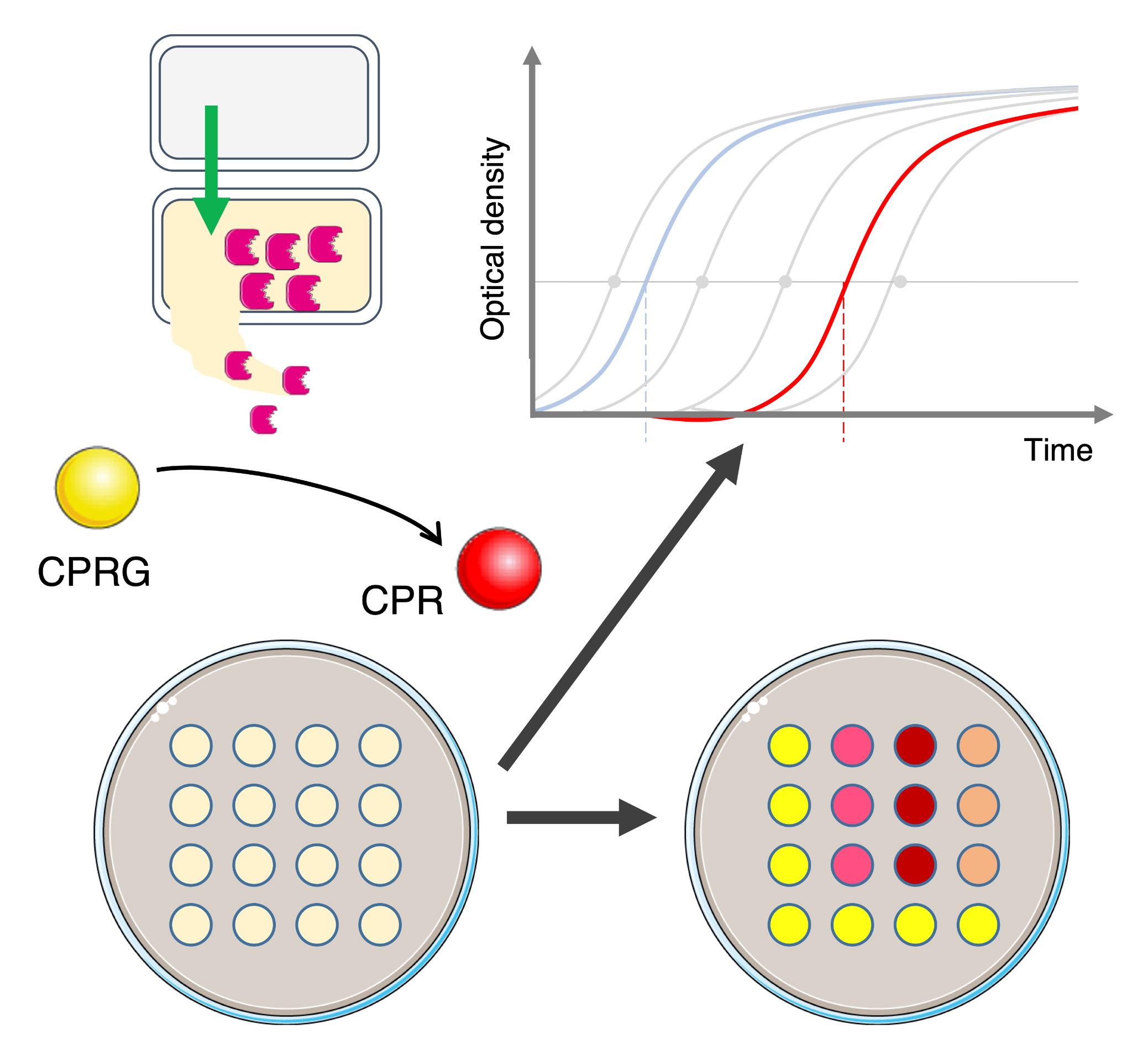
Evaluating bacterial competition in laboratory conditions often requires mixing attacker and recipient cells and, after co-incubation, counting attacker and surviving recipient cell colony-forming units on selective solid growth medium (MacIntyre et al., 2010; Flaugnatti et al., 2016; Alcoforado Diniz et al., 2017). However, this method is labor intensive, low throughput, costly, and time consuming. There is, therefore, a need to overcome these limitations, particularly when a large number of strains needs to be tested or in the case of a natural antibiotic resistant model that does not allow recovering prey cells in selective medium (Hazan et al., 2012; Lin and Lai, 2020). Recently, a new procedure based on the cell-impermeable β-galactosidase chromogenic substrate chlorophenol-red β-D-galactopyranoside (CPRG) was proposed (Vettiger and Basler, 2016). Because of its inability to cross the cell envelope, the CPRG (yellow) is only hydrolyzed in chlorophenol red (CPR, red) when the cytoplasmic β-galactosidase is released from a lacZ+ reporter cell after lysis. In addition to the visual evaluation of the antibacterial activity, the colorimetric properties of CPR can be measured by absorbance at λ = 572 nm (A572) or fluorescence (excitation at λ = 580 nm and emission at λ = 620 nm, F620nm) for a semi-quantitative value (Sicard et al., 2014; Vettiger and Basler, 2016; Figure 1A).
Here, this colorimetric method, called LAGA for Lysis-associated β-galactosidase Assay, was optimized to evaluate the activity of various antibacterial mechanisms. In addition, we developed a quantitative and complementary method based on growth recovery of surviving cells, called SGK for Survivors Growth Kinetics, to measure the number of surviving reporter cells after a competition assay, which is directly linked to the antibacterial activity of the attacker cell (Figure 1B). This method measures the time necessary for a subculture of recipient cells in selective medium to reach a defined optical density at λ = 600 nm (A600) after a competition assay, which is dependent on the initial number of surviving cells. By using standard curves from different known reporter inoculums, the initial number of surviving recipients can be estimated. Here, we describe the application of these methods in different bacterial models using various antibacterial strategies.
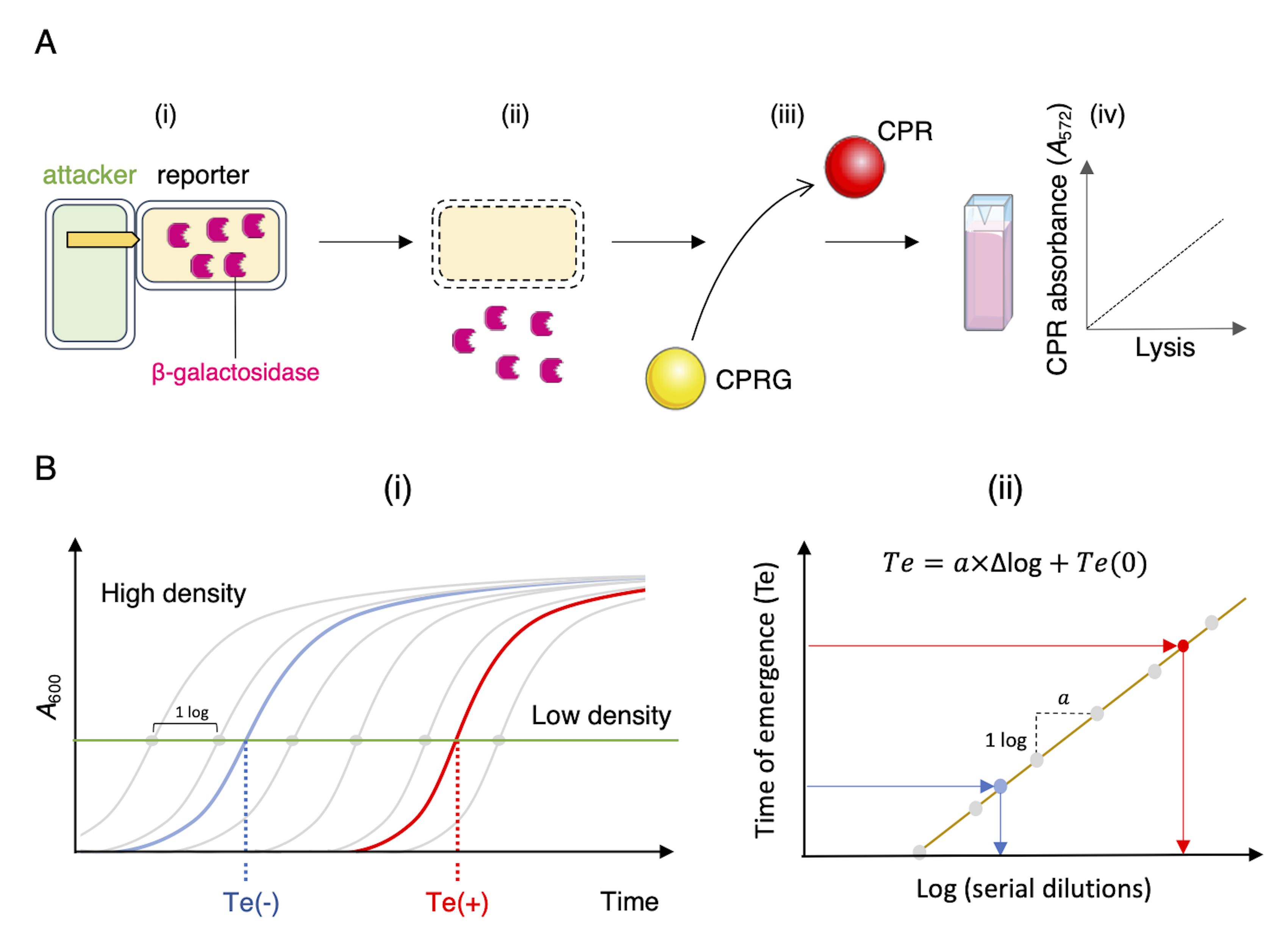
Figure 1. Schematic representations of the LAGA and SGK assays. (A) Lysis-associated β-galactosidase assay (LAGA). (i) Attacker and lacZ+ reporter cells are mixed. (ii) The antibacterial activity of the attacker results in reporter cell lysis and release of β-galactosidase to the medium. (iii) The hydrolysis of the yellow-colored chlorophenol-red β-D-galactopyranoside (CPRG) into the purple product chlorophenol red (CPR) by the released β-galactosidase can be measured spectrophotometrically (iv). The intensity of the coloration is correlated with CPR concentration and thus directly proportional to the number of lysed reporter cells and to the attacker antibacterial activity. (B) Survivors growth kinetics (SGK). (i) After mixing attacker and reporter cells and incubation, antibiotic-resistant reporter cells are selected on liquid medium containing the antibiotic. The growth curve is compared to standard growth curves of a serially diluted reporter culture (grey lines and dots). A high-density sample (no competition, blue line) will thus reach a defined A600 (horizontal green line) faster than the low-density sample (active competition, red line). The time of emergence [Te, time necessary to reach the defined A600; Te(-), no competition; Te(+), active competition] can be plotted to the linear regression curve of slope α, obtained with the serially-diluted standards (ii), allowing the evaluation of the initial number of surviving cells in the sample.
Materials and reagents
Chlorophenol red β-D-galactopyranoside (CPRG), sodium salt (Roche, catalog number: 10884308001; Sigma-Aldrich, catalog number: 220588; or equivalent). Store powder and soluble preparation (1.2 mg/mL in water) at -20 °C for up to one year or at 4 °C for up to one week
Non-treated, clear, flat-bottom 96-well microtiter plates (Greiner Cellstar®, catalog number: M0812; Nunc, catalog number: 266120; or equivalent)
Equipment
Spectrophotometer for reading absorbance at λ = 572 and 600 nm
Microplate reader (Tecan, Infinite 200; or comparable microplate reader)
Incubators to grow bacterial cultures and to incubate plates.
1.5- and 2-mL plastic tubes
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Taillefer, B., Grandjean, M. M., Herrou, J., Robert, D., Mignot, T., Sebban-Kreuzer, C. and Cascales, E. (2023). Qualitative and Quantitative Methods to Measure Antibacterial Activity Resulting from Bacterial Competition. Bio-protocol 13(13): e4706. DOI: 10.21769/BioProtoc.4706.
分类
微生物学 > 抗微生物试验 > 杀伤试验
细胞生物学 > 细胞活力 > 细胞裂解
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












