- EN - English
- CN - 中文
Autolysin Production from Chlamydomonas reinhardtii
莱茵衣藻自溶素的生产
发布: 2023年07月05日第13卷第13期 DOI: 10.21769/BioProtoc.4705 浏览次数: 1690
评审: Ansul LokdarshiSamed DelicNoelia Foresi

相关实验方案

利用SP3珠和稳定同位素质谱技术优化蛋白质合成速率:植物核糖体的案例研究
Dione Gentry-Torfer [...] Federico Martinez-Seidel
2024年05月05日 2870 阅读

基于活性蛋白质组学和二维聚丙烯酰胺凝胶电泳(2D-PAGE)鉴定拟南芥细胞间隙液中的靶蛋白酶
Sayaka Matsui and Yoshikatsu Matsubayashi
2025年03月05日 1990 阅读
Abstract
Chlamydomonas reinhardtii is a model organism for various processes, from photosynthesis to cilia biogenesis, and a great chassis to learn more about biofuel production. This is due to the width of molecular tools available, which have recently expanded with the development of a modular cloning system but, most importantly, with CRISPR/Cas9 editing now being possible. This technique has proven to be more efficient in the absence of a cell wall by using specific mutants or by digesting Chlamydomonas cell wall using the mating-specific metalloprotease autolysin (also called gametolysin). Multiple protocols have been used and shared for autolysin production from Chlamydomonas cells; however, they provide very inconsistent results, which hinders the capacity to routinely perform CRISPR mutagenesis. Here, we propose a simple protocol for autolysin production requiring transfer of cells from plates into a dense liquid suspension, gametogenesis by overnight incubation before mixing of gametes, and enzyme harvesting after 2 h. This protocol has shown to be highly efficient for autolysin production regardless of precise control over cell density at any step. Requiring a minimal amount of labor, it will provide a simple, ready-to-go approach to produce an enzyme critical for the generation of targeted mutants.
Graphical overview
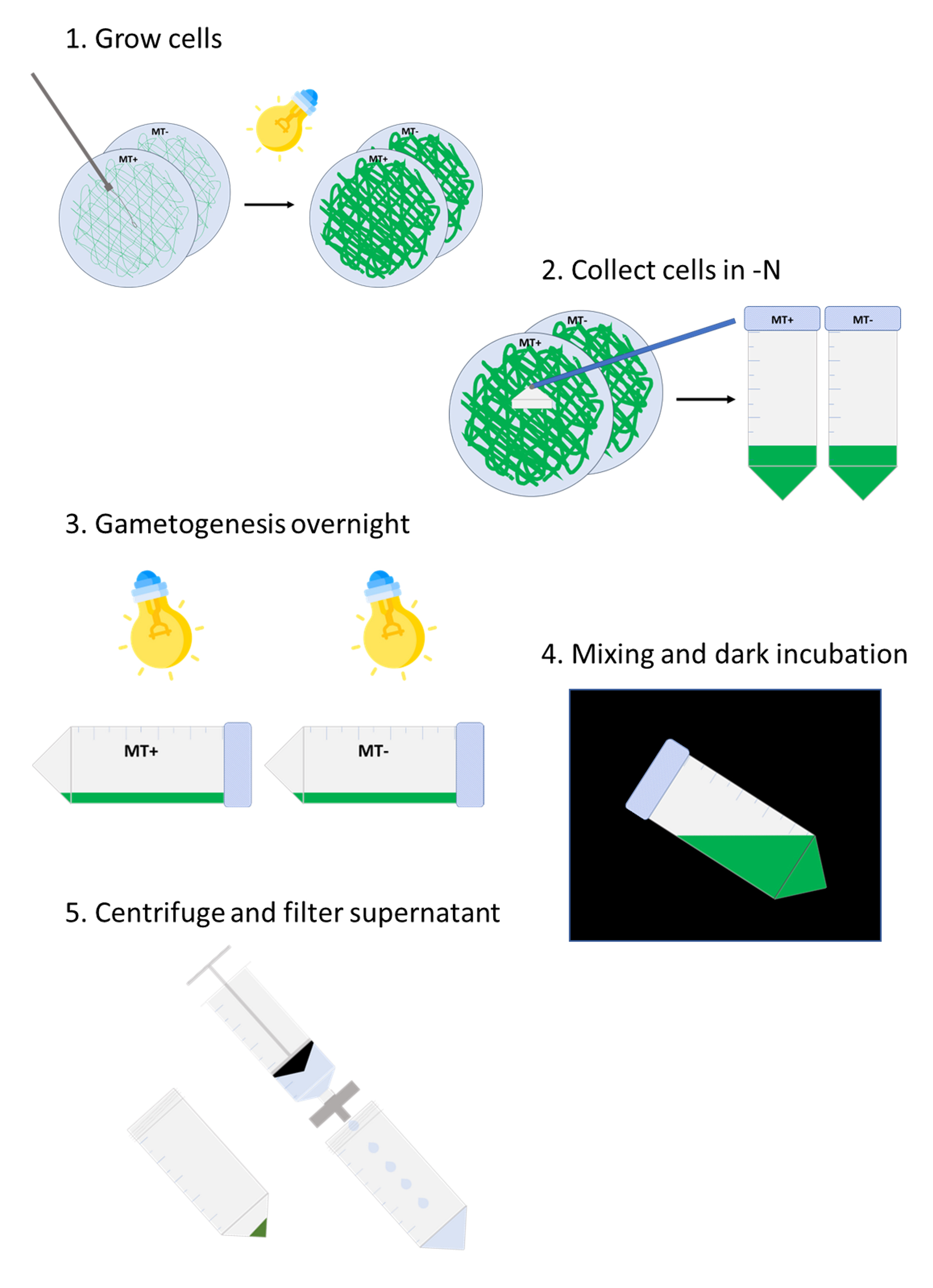
Workflow for autolysin production from Chlamydomonas reinhardtii
Background
Chlamydomonas’ genetic transformation has been possible for random insertional mutagenesis for over three decades (Kindle, 1990). In the last six years, methods have been developed for the precise editing of Chlamydomonas genome using the CRISPR technology by transfection of the ribonucleoprotein complex (RNP) directly through electroporation (Shin et al., 2016). Results have made clear that the cell wall is a strong barrier to entry of the RNP, as editing efficiency is usually much higher in cell wall–less strains or after digestion of the cell wall. Cell wall degradation can be performed using a commercially available enzyme called subtilisin/alcalase, but it has been shown to be only half as efficient as autolysin (Hwang et al., 2018), an enzyme produced by Chlamydomonas during mating in order to digest the cell wall and allow cell fusion of the two partners (Matsuda and Kubo, 2004). Autolysin can be extracted from a mixture of mating cells by centrifugation, preserved by freezing at -80 °C, and used later on any given walled strain to prepare for transformation. Several protocols are available in the literature as well as on the Chlamydomonas Resource Center website, but their efficiency has proven inconsistent in our hands. Here, we present a very simple protocol that requires minimal monitoring of cell growth and density, with overall > 70% efficiency. This will allow researchers interested in using the CRISPR technology to produce good quality autolysin and obtain mutated strains in their gene of interest, regardless of their favorite wildtype strain.
Materials and reagents
90 mm Petri dishes
Inoculating loop
Sterile 50 mL centrifuge tubes
Cell scraper
Pipette tips
1.7 mL microcentrifuge tubes
50 mL conical tubes
50 mL Luer-lock syringe
0.45 μm PES filter (VWR, catalog number: 76479-020)
Permanent marker pen
Microscope slides with eight wells (75 mm × 25 mm) (MP Biomedicals, catalog number: 096040805)
Plastic spectrophotometer cuvette (VWR, catalog number: 97000-586)
1 L filtration unit (VWR, catalog number: 10040-440)
Mating type (MT)+ wildtype strain CC-125 (available from the Chlamydomonas Resource Center, www.chlamy.org)
MT- wildtype strain CC-124 (available from the Chlamydomonas Resource Center, www.chlamy.org)
Note: Other pairs of MT+/MT- strains can be used; CC-620/621 are usually recommended due to their high mating efficiency.
Cell wall–less wildtype strain CC-503 (cw92, available from the Chlamydomonas Resource Center, www.chlamy.org)
Ammonium chloride (NH4Cl) (Sigma, catalog number: A9434)
Sodium chloride (NaCl) (Sigma, catalog number: 746398)
Calcium chloride dihydrate (CaCl2·2H2O) (Sigma, catalog number: C3881)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma, catalog number: 63140)
Tris base (J.T. Baker, catalog number: 4099-07)
Potassium phosphate monobasic (KH2PO4) (J.T. Baker, catalog number: 3246-07)
Potassium phosphate dibasic (K2HPO4) (J.T. Baker, catalog number: 3252-07)
Ethylenediaminetetraacetic acid disodium salt dihydrate (Na2EDTA·2H2O) (Sigma, catalog number: E5134)
Iron (II) sulfate heptahydrate (FeSO4·7H2O) (Sigma, catalog number: F-8048)
Zinc sulfate heptahydrate (ZnSO4·7H2O) (Sigma, catalog number: Z0251)
Boric acid (H3BO3) (Sigma, catalog number: B6768)
Manganese (II) chloride tetrahydrate (MnCl2·4H2O) (Sigma, catalog number: 221279)
Copper (II) chloride dihydrate (CuCl2·2H2O) (Sigma, catalog number: 467847)
Sodium molybdate dihydrate (Na2MoO4·2H2O) (J.T. Baker, catalog number: 3764)
Cobalt (II) chloride hexahydrate (CoCl2·6H2O) (Sigma, catalog number: C2644)
Glacial acetic acid (Sigma, catalog number: 320099)
Triton X-100 (Sigma, catalog number: T-9284)
Granulated agar (Fisher Scientific, catalog number: BP9744-5)
0.1% Triton X-100 (see Recipes)
Sucrose (Sigma, catalog number: S0389)
Stock solutions
Salts +N solution (20×) (see Recipes)
Salts -N solution (20×) (see Recipes)
2 M Tris Base (100×)
Phosphate buffer (200×) (see Recipes)
Hutner’s trace elements (200×) (see Recipes, also available from the Chlamydomonas Resource Center, www.chlamy.org)
TAP (Tris-Acetate-Phosphate) liquid medium (see Recipes)
TAP 1.5% agar medium (see Recipes)
TAP -N liquid medium (see Recipes)
TAP sucrose 40 mM (see Recipes)
Equipment
Pipettes
Centrifuge and microcentrifuge
Light microscope
Laminar flow hood
Glass beaker
Spectrophotometer
Vacuum pump
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Findinier, J. (2023). Autolysin Production from Chlamydomonas reinhardtii. Bio-protocol 13(13): e4705. DOI: 10.21769/BioProtoc.4705.
分类
植物科学 > 植物分子生物学 > DNA > DNA 修饰
植物科学 > 植物生物化学 > 蛋白质 > 分离和纯化
生物化学 > 蛋白质 > 分离和纯化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link









