- EN - English
- CN - 中文
Revised iCLIP-seq Protocol for Profiling RNA–protein Interaction Sites at Individual Nucleotide Resolution in Living Cells
用于在活细胞中以单个核苷酸分辨率分析RNA-蛋白质相互作用位点的修订iCLIP-seq方案
发布: 2023年06月05日第13卷第11期 DOI: 10.21769/BioProtoc.4688 浏览次数: 5225
评审: Gal HaimovichMarion HoggAnonymous reviewer(s)
Abstract
Individual nucleotide resolution UV cross-linking and immunoprecipitation followed by high-throughput sequencing (iCLIP-seq) is a powerful technique that is used to identify RNA-binding proteins’ (RBP) binding sites on target RNAs and to characterize the molecular basis of posttranscriptional regulatory pathways. Several variants of CLIP have been developed to improve its efficiency and simplify the protocol [e.g., iCLIP2 and enhanced CLIP (eCLIP)]. We have recently reported that transcription factor SP1 functions in the regulation of alternative cleavage and polyadenylation through direct RNA binding. We utilized a modified iCLIP method to identify RNA-binding sites for SP1 and several of the cleavage and polyadenylation complex subunits, including CFIm25, CPSF7, CPSF100, CPSF2, and Fip1. Our revised protocol takes advantage of several features of the eCLIP procedure and also improves on certain steps of the original iCLIP method, including optimization of circularization of cDNA. Herein, we describe a step-by-step procedure for our revised iCLIP-seq protocol, that we designate as iCLIP-1.5, and provide alternative approaches for certain difficult-to-CLIP proteins.
Key features
• Identification of RNA-binding sites of RNA-binding proteins (RBPs) at nucleotide resolution.
• iCLIP-seq provides precise positional and quantitative information on the RNA-binding sites of RBPs in living cells.
• iCLIP facilitates the identification of sequence motifs recognized by RBPs.
• Allows quantitative analysis of genome-wide changes in protein-RNA interactions.
• Revised iCLIP-1.5 protocol is more efficient and highly robust; it provides higher coverage even for low-input samples.
Graphical overview
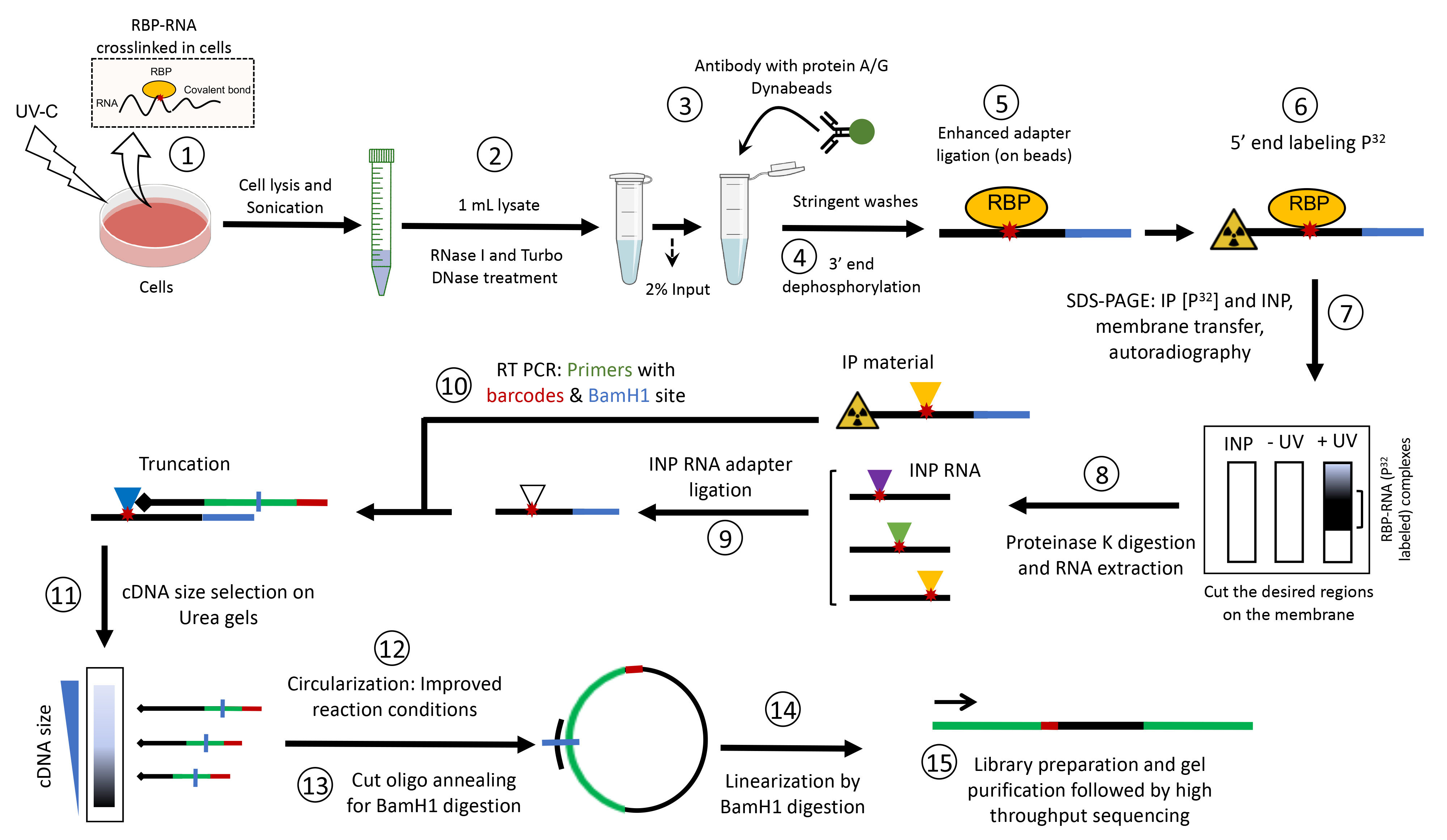
Background
RNA-binding proteins (RBPs) are key players in posttranscriptional regulation, as they regulate essentially all aspects of mRNA metabolism, including 5′ cap formation, pre-mRNA splicing, 3′ end formation, mRNA export, and mRNA stability (Hentze et al., 2018). RBPs that decorate an RNA molecule during its life can dynamically change in space and time while they tightly co-ordinate posttranscriptional regulation (Hafner et al., 2021). Increasingly, links are being detected between defects in RBPs’ functions and human diseases, including neurological disorders and cancer (Pereira et al., 2017). Thus, understanding the role of RBPs in posttranscriptional regulation, and identifying their full repertoire of RNA targets in cells, is of fundamental importance for both better characterizing gene expression regulatory programs and drug development.
Although numerous methods exist to characterize RBPs’ target RNAs in cells (e.g., RIP, TRIBE, and APEX-seq), the cross-linking and immunoprecipitation (CLIP)-based methods are considered the gold standard for precisely identifying endogenous RNA-binding sites of RBPs (Hafner et al., 2021). Several variants of CLIP have been developed, including HITS-CLIP, individual-nucleotide resolution CLIP (iCLIP), ir-CLIP, enhanced CLIP (eCLIP), and iCLIP2, aiming to simplify the protocol and improve its efficiency (Buchbender et al., 2020;Konig et al., 2011;Lee et al., 2021;Van Nostrand et al., 2016; Zarnegar et al., 2016). These methods all rely on exposure of live cells to UV-C light, which covalently crosslinks RBPs to their target RNAs. The CLIP method can be carried out so as to provide precise positional and quantitative information on the crosslink sites. In particular, iCLIP aims to detect RNA fragments that truncate at the crosslink site during cDNA preparation, and thus reveals RBP binding sites on target RNA at individual nucleotide resolution. Since the original iCLIP method is laborious, and the library preparation efficiency can become limiting for low input samples (Huppertz et al., 2014), eCLIP was developed to omit the inefficient cDNA circularization step, improve the adapter ligation conditions, and implement a size-matched input (SMI) to control for background signal (Van Nostrand et al., 2016). Although eCLIP maintains the individual nucleotide resolution of iCLIP, it does not include direct visualization of protein-associated RNA. Hence, the quality of immunoprecipitated RNA, as well as the possibility of co-purifying background RBPs, cannot be directly monitored.
We have recently modified the original iCLIP method to interrogate the posttranscriptional regulatory functions of the ubiquitously expressed transcription factor SP1 (Song et al., 2022). By identifying its RNA-binding sites, we found that SP1 preferentially binds to A/G-rich sequence motifs on target RNAs. We further utilized our revised iCLIP procedure to examine the binding profiles of several cleavage and polyadenylation factors on target mRNAs (Song et al., 2022). Our data yielded high-resolution RNA-binding maps for SP1, as well as the cleavage and polyadenylation complex subunits, and unraveled a previously unknown function of SP1 in alternative cleavage and polyadenylation regulation. We have used this protocol with several cell lines (e.g., mouse neuroblastoma N2a cells, cgr8 embryonic stem cells, HeLa cells, glioblastoma, and HEK293 cells) to identify RNA targets of RBPs involved in diverse functions, including splicing regulation, mRNA stability, mRNA export, 5′ capping, and mRNA modifications (Nitoiu et al., 2021;Han et al., 2017and 2022;Nabeel-Shah et al., 2022 ). Our revised iCLIP protocol combines the improvements previously implemented in eCLIP (e.g., enhanced adapter ligation and SMI), with the optimization of several steps in the original iCLIP method, including improved circularization of the cDNA by increasing the incubation time and adding Betain in the reaction mixture, which helps the circularization of difficult-to-ligate substrates. Moreover, we utilize two-step dephosphorylation of RNA fragments to ensure the complete removal of the 3′-cyclic phosphate group left behind by RNase I cleavage. Since iCLIP2 is more similar to eCLIP (i.e., omission of circularization and linearization steps), whereas our protocol essentially follows the workflow of the original iCLIP procedure, we designate our revised protocol as iCLIP-1.5.
Materials and reagents
Protein G Dynabeads (Life Technologies, catalog number: 10004D)
Protein A Dynabeads (Life Technologies, catalog number: 10002D)
GFP recombinant rabbit monoclonal antibody (Thermo Fisher Scientific, catalog number: G10362)
Rabbit polyclonal anti-GFP (Abcam, catalog number: ab290)
SP1 antibody (Santa Cruz, catalog number: sc-17824)
Protease Inhibitor Cocktail Set III (Calbiochem/Merck, catalog number: 539134-1SET)
RNase I (Life Technologies, catalog number: AM2295)
Turbo DNase (Life Technologies, catalog number: AM2238)
T4 PNK plus 10× PNK buffer (NEB, catalog number: M0201L)
Murine RNase inhibitor 40 U/µL (NEB, catalog number: M0314L)
T4 RNA ligase 1 high conc 30 U/µL (NEB, catalog number: M0437M)
10× T4 RNA ligase reaction buffer (NEB, catalog number: B0216SVIAL)
FastAP 1 U/µL (Life Technologies, catalog number: EF0652)
Proteinase K 0.8 U/µL (NEB, catalog number: P8107S)
MyONE Silane beads (Life Technologies, catalog number: 37002D)
Pre-adenylated adapter L3-App IDT (rAppAGATCGGAAGAGCGGTTCAG/ddC/)
ATP [γ-32P] (PerkinElmer, catalog number: NEG502A250UC)
4%–12% NuPage gels (Life Technologies, catalog number: NP0322BOX)
LDS-4× sample buffer (NuPage loading buffer) (Life Technologies, catalog number: NP0007)
Pre-stained protein size marker (Fermentas, catalog number: SM1811)
Nitrocellulose membrane protran BA85 (VWR, catalog number: 732-4174)
20× transfer buffer (Life Technologies, catalog number: NP0006-1)
20× MOPS-SDS running buffer (Life Technologies, catalog number: NP0001)
Whatman filter paper (GE Healthcare, catalog number: 3030917)
Film (Fuji, catalog number: 4741019236)
Phenol/chloroform (Sigma, catalog number: P3803)
Phase lock gel heavy tube (VWR, catalog number: 713-2536)
GlycoBlue (Ambion, catalog number: 9510)
3 M sodium acetate pH 5.5 (Life Technologies, catalog number: AM9740)
2× TBE-urea loading buffer (Life Technologies, catalog number: LC6876)
6% TBE-urea pre-cast gels (Life Technologies, catalog number: EC68652B)
Low molecular weight marker (NEB, catalog number: N3233L)
TBE running buffer (Life Technologies, catalog number: LC6675)
SYBR green II (Life Technologies, catalog number: S-7564)
19 G syringe needle (BD, Microlance, catalog number: 300637)
Glass pre-filters (Whatman, catalog number: 1823010)
Costar SpinX column (Corning, catalog number: 8161)
CircLigaseTM (Lucigen, catalog number: CL4115K)
10× Circligase buffer (Lucigen, catalog number: CL4115K)
Betain solution 5 M (Sigma, catalog number: B0300-1VL)
Cut_oligo ‘GTTCAGGATCCACGACGCTCTTCaaaa’ (HPLC-purified, IDT)
BamHI (Fermentas, catalog number: FD0055)
Fast digest buffer (Fermentas, catalog number: FD0055)
2× Phusion High-Fidelity PCR Master Mix (NEB, catalog number: M0531L)
NaOH (Merck/Sigma, catalog number: S8045-500G)
RLT buffer (Qiagen, catalog number: 79216)
PBS (Sigma-Aldrich, catalog number: D8537)
UltraPureTM agarose (Invitrogen, catalog number: 16500-500)
QIAquick Gel Extraction kit (Qiagen, catalog number: 28706)
Tris (VWR, catalog number: 0826)
Sodium chloride (NaCl) (VWR, catalog number: 27810.364)
Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M2670)
Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C1016)
Potassium chloride (KCl) (Thermo Fisher Scientific, catalog number: AM9640G)
Urea (Thermo Fisher Scientific, catalog number: 15505035)
Sodium deoxycholate (Thermo Fisher Scientific, catalog number: 89904)
Tween-20 (Thermo Fisher Scientific, catalog number: 28320)
Sodium dodecyl sulfate (SDS) (Thermo Fisher Scientific, catalog number: 28364)
Triton X-100 (Thermo Fisher Scientific, catalog number: 28314)
Primers (HPLC-purified from IDT):
P5_Solexa: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT
P3_Solexa: CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT
Rt1clip/5Phos/NNAACCNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt2clip/5Phos/NNACAANNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt3clip/5Phos/NNATTGNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt4clip/5Phos/NNAGGTNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt5clip/5Phos/NNCGCCNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt6clip/5Phos/NNCCGGNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt7clip/5Phos/NNCTAANNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt8clip/5Phos/NNCATTNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt9clip/5Phos/NNGCCANNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt10clip/5Phos/NNGACCNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt11clip/5Phos/NNGGTTNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt12clip/5Phos/NNGTGGNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt13clip/5Phos/NNTCCGNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt14clip/5Phos/NNTGCCNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt15clip/5Phos/NNTATTNNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Rt16clip/5Phos/NNTTAANNNAGATCGGAAGAGCGTCGTGgatcCTGAACCGC
Solutions
Lysis buffer (see Recipes)
High-salt wash (see Recipes)
PNK buffer (see Recipes)
PK buffer (see Recipes)
PK buffer + 7 M urea (see Recipes)
1× RNA ligase buffer (see Recipes)
10× RNA ligase buffer (see Recipes)
1× FastAP buffer (see Recipes)
Recipes
Lysis buffer
50 mM Tris-HCl, pH 7.4
100 mM NaCl
1% Igepal CA-630
0.1% SDS
0.5% sodium deoxycholate
On the day: 1/100 volume of Protease Inhibitor Cocktail Set III, for tissues: 1/1,000 volume of ANTI-RNase
High-salt wash
50 mM Tris-HCl, pH 7.4
1 M NaCl
1 mM EDTA
1% Igepal CA-630
0.1% SDS
0.5% sodium deoxycholate
PNK buffer
20 mM Tris-HCl, pH 7.4
10 mM MgCl2
0.2% Tween-20
PK buffer
100 mM Tris-HCl, pH 7.4
50 mM NaCl
10 mM EDTA
PK buffer + 7 M urea
100 mM Tris-HCl, pH 7.4
50 mM NaCl
10 mM EDTA
7 M urea
1× RNA ligase buffer (no DTT)
50 mM Tris-HCl pH 7.5
10 mM MgCl2
10× RNA ligase buffer (no DTT)
500 mM Tris-HCl pH 7.5
100 mM MgCl2
1× FastAP buffer
10 mM Tris pH 7.5
5 mM MgCl2
100 mM KCl
0.02% Triton X-100
Equipment
Refrigerated microcentrifuge
Picofuge (Thermo Fisher Scientific, catalog number: 75004061)
Stratalinker UV crosslinker 2400 Stratagene
Denaturing agarose gel apparatus (Bio-Rad Laboratories)
Magnetic stand (Thermo Fisher Scientific, Invitrogen)
Electrophoresis chamber (Life Technologies, catalog number: EI0002)
Transfer apparatus (Life Technologies, catalog number: EI0002)
Sponge pads for XCell II blotting (Life Technologies, catalog number: EI9052)
Thermoblock (Thermo Fisher Scientific, Invitrogen)
Thermomixer (Boekel Scientific)
Sonicator (Branson Digital Sonifier)
Geiger counter sensitive to beta particles (Ludlum 3 survey meter Geiger counter radiometer 44-9 pancake probe)
3/8" or 1/2" Plexiglas benchtop shield (P32 protective shield)
Cell scraper
Liquid nitrogen (N2)
Saran wrap
Blue light transilluminator
UV transilluminator
X-ray film cassette
Software
iMAPs (Jernej Ule’s lab, https://imaps.goodwright.com/)
PureCLIP (Sabrina Krakau, https://pureclip.readthedocs.io/en/latest/index.html)
CTK package (Chaolin Zhang’s lab, https://zhanglab.c2b2.columbia.edu/index.php/CTK_Documentation)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Nabeel-Shah, S. and Greenblatt, J. F. (2023). Revised iCLIP-seq Protocol for Profiling RNA–protein Interaction Sites at Individual Nucleotide Resolution in Living Cells. Bio-protocol 13(11): e4688. DOI: 10.21769/BioProtoc.4688.
分类
分子生物学 > DNA > DNA-蛋白质相互作用
系统生物学 > 基因组学 > 测序
生物化学 > RNA > RNA-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













