- EN - English
- CN - 中文
Detecting Photoactivatable Cre-mediated Gene Deletion Efficiency in Escherichia coli
检测大肠杆菌中光激活的Cre介导的基因缺失效率
发布: 2023年06月05日第13卷第11期 DOI: 10.21769/BioProtoc.4685 浏览次数: 1943
评审: Chiara AmbrogioMatthew SwireAdriano BolondiAnonymous reviewer(s)

相关实验方案

将Miniprep制备的大肠杆菌K12菌株质粒DNA转化为可用于植物遗传转化的根癌农杆菌EHA105细胞的简单可靠方法
Beenzu Siamalube [...] Steven Runo
2025年01月05日 2373 阅读
Abstract
Gene deletion is one of the standard approaches in genetics to investigate the roles and functions of target genes. However, the influence of gene deletion on cellular phenotypes is usually analyzed sometime after the gene deletion was introduced. Such lags from gene deletion to phenotype evaluation could select only the fittest fraction of gene-deleted cells and hinder the detection of potentially diverse phenotypic consequences. Therefore, dynamic aspects of gene deletion, such as real-time propagation and compensation of deletion effects on cellular phenotypes, still need to be explored. To resolve this issue, we have recently introduced a new method that combines a photoactivatable Cre recombination system and microfluidic single-cell observation. This method enables us to induce gene deletion at desired timings in single bacterial cells and to monitor their dynamics for prolonged periods. Here, we detail the protocol for estimating the fractions of gene-deleted cells based on a batch-culture assay. The duration of blue light exposure significantly affects the fractions of gene-deleted cells. Therefore, gene-deleted and non-deleted cells can coexist in a cellular population by adjusting the duration of blue light exposure. Single-cell observations under such illumination conditions allow the comparison of temporal dynamics between gene-deleted and non-deleted cells and unravel phenotypic dynamics provoked by gene deletion.
Keywords: Gene deletion (基因缺失)Background
Analyzing the phenotypes of gene-deleted cell strains or multicellular organisms is the cornerstone of genetics studies. The functional roles of various genes in each organism can be deduced from the phenotypic changes caused by their deletion. For systematically analyzing the effects of gene deletion, gene knock-out libraries are available for several model organisms (Jorgensen et al., 2002; Baba et al., 2006; Kim et al., 2010; White et al., 2013). For example, the Keio collection (Baba et al., 2006) is a single-gene knock-out library of Escherichia coli that has been used to infer the roles of genes in various biological phenomena, such as cellular growth and morphogenesis (Campos et al., 2018), antibiotic sensitivity (Tamae et al., 2008), and evolution (Lukačišinová et al., 2020).
However, the significance of each gene might be context dependent. Gene expression levels change dynamically depending on environmental conditions (Schmidt et al., 2016; Bhatia et al., 2022). In Koganezawa et al. (2022), we induced a Cre-mediated antibiotic resistance gene deletion with blue-light illumination in the presence of the antibiotic. The microfluidic technique revealed that a small fraction of the gene-deleted cells could continue their growth without other genetic changes, and the fraction varied depending on the history of antibiotic administration. These results indicate that the impact of gene deletion on cellular physiology can depend on historical conditions (Koganezawa et al., 2022). Therefore, revealing dynamic genotype–phenotype relationships is crucial to understand the functional roles of each gene in given biological contexts.
Optogenetic techniques are useful for analyzing dynamic genotype–phenotype relationships. In particular, the photoactivatable Cre (PA-Cre) system allows us to induce gene deletion at desired timings by blue light exposure (Kawano et al., 2016). The PA-Cre system consists of split Cre recombinase, CreC, and CreN, fused to p-Magnet (p-Mag) and n-Magnet (n-Mag) protein fragments, respectively (Kawano et al., 2016). p-Mag and n-Mag are derived from a fungal photoreceptor Vivid and form a dimer upon blue light illumination (Kawano et al., 2015). Therefore, when these components of the PA-Cre system are expressed in the cytoplasm of bacterial cells, blue light illumination can induce the deletion of a target gene sandwiched between the two loxP sequences at desired timings. We constructed E. coli cell strains that harbor the PA-Cre system and the photocleavable fluorescently labeled chloramphenicol-resistance gene (Koganezawa et al., 2022). We observed the E. coli cells at the single-cell level in the microfluidic device and deleted the genes directly in the device by blue light illumination.
The duration of blue light illumination significantly influences the fraction of gene-deleted cells in the population. Therefore, by adjusting the illumination duration, we can simultaneously observe the dynamics of gene-deleted cells and non-deleted cells in a cell population. Since the target chloramphenicol resistance gene is tagged with the gene for the fluorescent protein mCherry, the fractions of gene-deleted cells are measurable simply by fluorescent microscopy. Here, we describe the protocol for measuring the fraction of gene-deleted cells based on batch-culture assay.
Materials and reagents
Glass test tube ϕ 16.5 × 165 mm (IWAKI, catalog number: B14-002-160)
Molton cap (AS ONE, catalog number: 6-352-07)
1.5 mL microtube (TreffLab, catalog number: 96.07246.9.01)
Minisart® syringe filter (Sartorius, catalog number: S7597-FXOSK)
Sterilized Petri dish ϕ90 × 15 mm (AS ONE, catalog number: 1-7484-01)
Aluminum foil
E. coli strain, carrying a fluorescently tagged target gene flanked with two loxP sequences and photoactivatable Cre (PA-Cre) system. Here, we used YK0083, which has a chloramphenicol resistance gene, cat, as a target gene for demonstration. The mCherry-tagged cat gene sandwiched with two loxP sequences was introduced into the intC locus on the genome. The genes for the PA-Cre system, creN-nmag and pmag-creC, are placed downstream of PLlacO1 promoter on a plasmid. These genes are inducible by IPTG. This strain is available by contacting the authors.
DifcoTM LB broth, Miller (Luria-Bertani) (BD, catalog number: 244620)
Agar (Wako, catalog number: 010-15815)
DifcoTM M9 minimal salts, 5× (BD, catalog number: 248510)
D(+)-glucose (Wako, catalog number: 049-31165)
MgSO4·7H2O (Wako, catalog number: 131-00405)
CaCl2·2H2O (Wako, catalog number: 31-00435)
MEM amino acids solution (50×) (SIGMA, catalog number: M5550)
Ampicillin sodium (Wako, catalog number: 016-23301)
Isopropyl-β-D(-)-thiogalactopyranoside (IPTG) (Wako, catalog number: 094-05144)
Elix® water produced by Milli-Q® Integral 3 (Merck, catalog number: ZRXQ003T0)
Milli-Q® water produced by Milli-Q® Integral 3 (Merck, catalog number: ZRXQ003T0)
LB broth (see Recipes)
LB agar containing 100 μg/mL of ampicillin (Amp) (see Recipes)
M9 medium with 0.2% glucose and amino acids (see Recipes)
1 M IPTG (see Recipes)
50 mg/mL Amp (see Recipes)
Equipment
Centrifuge (Hitachi, himac, CT 15RE)
Spectrometer (Shimadzu, UV-1800)
Blue light illuminator (Power Supply: CCS Inc., ISC-201-2; Blue light Illuminator: CCS Inc., SLM-150X150-BB)
BioShaker (TAITEC, BR-21FP)
Incubator (MITSUBISHI ELECTRIC ENGINEERING, SLC-25A)
Stereomicroscope (Stereomicroscope: Olympus, SZ61; LED source: NIGHTSEA, SFA-GR)
Portable photodiode-based laser power meter (Gentec-EO, PRONTO-Si)
Procedure
Sample preparation
Culture E. coli cells in LB broth with 50 μg/mL of Amp at 37 °C with shaking overnight.
Centrifuge 100 μL of the overnight culture at 21,500× g for 1 min at room temperature.
Remove the supernatant.
Resuspend the cellular pellet in 1 mL of M9 medium with 0.2% glucose and amino acids.
Measure the OD600 of the resuspended cells.
Adjust the OD600 to 0.001 in 2 mL of M9 medium with 0.2% glucose and amino acids containing 50 μg/mL of Amp and 0.1 mM IPTG.
Cover the test tube with the aluminum foil.
Cultivate the cells at 37 °C with shaking for 3 h.
Gene deletion induction
Remove the aluminum foil after the cultivation.
Expose the test tube to blue light using blue-light illuminator (light intensity: 6.8 mW) for the desired duration (Figures 1 and 2, see Notes).

Figure 1. Blue-light illumination inside a bio shaker with an illuminator. A blue-light illuminator was directly fastened onto a test tube spring holder inside a bio shaker. It casts a blue light downward on E. coli cultures. Light intensity was adjusted to 6.8 mW, measured with a laser power meter (λ = 470 nm).
Calculation of gene-deleted cells in batch
Dilute the cultures exposed to blue light to OD600 = 1.0 × 10-6, which is equivalent to approximately 7 × 102 cells/mL.
Spread 150 μL of the diluted cultures on LB agar plate containing 100 μg/mL of Amp.
Cover the agar plates with aluminum foil.
Incubate the plates at 37 °C for 18 h.
Count the number of colonies under ambient light and excitation light for examining mCherry fluorescence using a stereomicroscope (Figure 2A).
Calculate the fraction of gene-deleted cells as the number of non-fluorescent colonies divided by the number of total colonies.
Notes
The fraction of gene-deleted cells varies depending on the duration of blue-light illumination (Figure 2B). Therefore, an experimenter should determine the duration of blue-light exposure depending on the purpose of the experiment.
Cre-mediated gene deletion with blue-light illumination is detectable with loss of mCherry fluorescent signal.
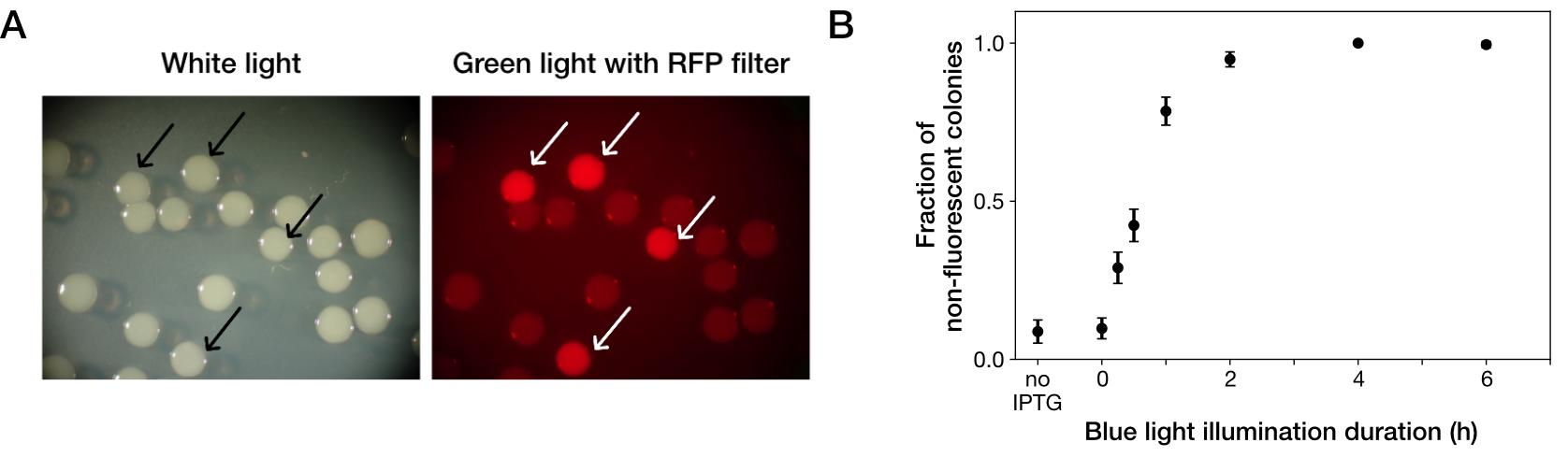
Figure 2. Photoactivatable Cre-mediated gene deletion. (A) Stereomicroscopic pictures of YK0083 colonies after blue-light illumination under ambient light (left) and excitation light for detecting mCherry fluorescence (right). Cells were exposed to blue light for 1 h. Arrows show colonies generated from cells from which target genes were not deleted with blue-light illumination. (B) Blue-light illumination duration dependency of gene deletion efficiency. In case of less than 6 h blue-light illumination, test tubes were covered with aluminum foil and shaken at 37 °C, so that the time from the start of blue-light illumination to spreading on the LB agar was 6 h and consistent among the conditions. “no IPTG” indicates the condition where cells were cultured without IPTG and not exposed to blue light. Black points indicate the means. Error bars indicate standard errors (N = 239 for no IPTG, N = 327 for 0 h, N = 335 for 0.25 h, N = 373 for 0.5 h, N = 344 for 1 h, N = 350 for 2 h, N = 296 for 4 h, and N = 209 for 6 h). Reprinted/adapted from Koganezawa et al. (2022).
Recipes
LB broth
Mix the following:
25 g of DifcoTM LB broth, Miller (Luria-Bertani)
1 L of Elix® water
Autoclave at 121 °C for 15 min.
Store at room temperature.
At the time of use, add 50 mg/mL Amp on a clean bench as appropriate, if needed.
LB agar containing 100 μg/mL of Amp
Mix the following:
25 g of DifcoTM LB broth, Miller (Luria-Bertani)
15 g of agar
1 L of Elix® water
Autoclave at 121 °C for 15 min.
Add 2 mL of 50 mg/mL Amp on a clean bench.
Dispense 25 mL in each 90 mm dish on a clean bench.
Store at 4 °C.
M9 medium with 0.2% glucose and amino acids
Prepare the following reagents:
5× M9
1) Dissolve 56.4 g of DifcoTM M9 minimal salts, 5× in 1 L of Milli-Q® water.
2) Autoclave at 121 °C for 15 min.
3) Store at room temperature.
20% glucose
1) Dissolve 10 g of D(+)-glucose in Milli-Q® water to 50 mL.
2) Sterilize using a 0.2 μm pore filter.
3) Store at 4 °C.
1 M MgSO4
1) Dissolve 12.3 g of MgSO4·7H2O in Milli-Q® water to 50 mL.
2) Sterilize using a 0.2 μm pore filter.
3) Store at 4 °C.
1 M CaCl2
1) Dissolve 7.4 g of CaCl2·2H2O in Milli-Q® water to 50 mL.
2) Sterilize using a 0.2 μm pore filter.
3) Store at 4 °C.
Sterilized water
1) Autoclave Milli-Q® water at 121 °C for 15 min.
2) Store at room temperature.
Mix the following on a clean bench:
200 mL of 5× M9
10 mL of 20% glucose
10 mL of MEM amino acids solution (50×)
2 mL of 1 M MgSO4
100 μL of 1 M CaCl2
778 mL of sterilized water
Store at 4 °C.
At the time of use, add antibiotics or IPTG on a clean bench as appropriate, if needed.
1 M IPTG
Dissolve 2.383 g of IPTG in Milli-Q® water to 10 mL.
Sterilize using a 0.2 μm pore filter.
Dispense in microtubes on a clean bench.
Store dispensed tubes at -20 °C.
50 mg/mL Amp
Dissolve 500 mg of ampicillin sodium in 10 mL of Milli-Q® water.
Sterilize using a 0.2 μm pore filter.
Dispense in microtubes on a clean bench.
Store dispensed tubes at -20 °C.
Acknowledgments
This work was supported by JST CREST Grant Number JPMJCR1927 (Y.W.) and JPMJCR1653 (M.S.); JST ERATO Grant Number JPMJER1902 (Y.W.); Japan Society for the Promotion of Science KAKENHI Grant Number 17H06389 and 19H03216 (Y.W.); Project Grant from Kanagawa Institute of Industrial Science and Technology (KISTEC) (M. S.); and Grant-in-Aid for JSPS Fellows Grant Number JP19J22506 (Y.K.). This protocol was derived from Koganezawa et al. (2022).
Competing interests
The authors declare that no competing interests exist.
References
- Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., Datsenko, K. A., Tomita, M., Wanner, B. L. and Mori, H. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006 0008.
- Bhatia, R. P., Kirit, H. A., Predeus, A. V. and Bollback, J. P. (2022). Transcriptomic profiling of Escherichia coli K-12 in response to a compendium of stressors. Sci Rep 12(1): 8788.
- Campos, M., Govers, S. K., Irnov, I., Dobihal, G. S., Cornet, F. and Jacobs-Wagner, C. (2018). Genomewide phenotypic analysis of growth, cell morphogenesis, and cell cycle events in Escherichia coli. Mol Syst Biol 14(6): e7573.
- Jorgensen, P., Nishikawa, J. L., Breitkreutz, B. J., and Tyers, M. (2002). Systematic identification of pathways that couple cell growth and division in yeast. Science 297(5580): 395-400.
- Kawano, F., Okazaki, R., Yazawa, M., and Sato, M. (2016). A photoactivatable Cre–loxP recombination system for optogenetic genome engineering. Nat Chem Biol 12(12): 1059-1064.
- Kawano, F., Suzuki, H., Furuya, A. and Sato, M. (2015). Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun 6: 6256.
- Kim, D. U., Hayles, J., Kim, D., Wood, V., Park, H. O., Won, M., Yoo, H. S., Duhig, T., Nam, M., Palmer, G., et al. (2010). Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol, 28(6): 617-623.
- Koganezawa, Y., Umetani, M., Sato, M., and Wakamoto, Y. (2022). History-dependent physiological adaptation to lethal genetic modification under antibiotic exposure. eLife 11: e74486.
- Lukačišinová, M., Fernando, B., and Bollenbach, T. (2020). Highly parallel lab evolution reveals that epistasis can curb the evolution of antibiotic resistance. Nat Commun 11(1): 3105.
- Schmidt, A., Kochanowski, K., Vedelaar, S., Ahrne, E., Volkmer, B., Callipo, L., Knoops, K., Bauer, M., Aebersold, R. and Heinemann, M. (2016). The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol 34(1): 104-110.
- Tamae, C., Liu, A., Kim, K., Sitz, D., Hong, J., Becket, E., Bui, A., Solaimani, P., Tran, K. P., Yang, H., et al. (2008). Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J Bacteriol, 190(17): 5981–5988.
- White, J. K., Gerdin, A. K., Karp, N. A., Ryder, E., Buljan, M., Bussell, J. N., Salisbury, J., Clare, S., Ingham, N. J., Podrini, C., et al. (2013). Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154(2): 452-464.
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Koganezawa, Y., Wakamoto, Y., Sato, M. and Umetani, M. (2023). Detecting Photoactivatable Cre-mediated Gene Deletion Efficiency in Escherichia coli. Bio-protocol 13(11): e4685. DOI: 10.21769/BioProtoc.4685.
- Koganezawa, Y., Umetani, M., Sato, M., and Wakamoto, Y. (2022). History-dependent physiological adaptation to lethal genetic modification under antibiotic exposure. eLife 11: e74486.
分类
微生物学 > 微生物遗传学
微生物学 > 微生物生理学 > 适应
生物科学 > 生物技术 > 微生物技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











