- EN - English
- CN - 中文
A Miniature Sucrose Gradient for Polysome Profiling
用于多聚核糖体分析的微型蔗糖梯度
发布: 2023年03月20日第13卷第6期 DOI: 10.21769/BioProtoc.4622 浏览次数: 4471
评审: Wenrong HeYao XiaoAnonymous reviewer(s)
Abstract
Polysome profiling by sucrose density gradient centrifugation is commonly used to study the overall degree of translation (messenger RNA to protein synthesis). Traditionally, the method begins with synthesis of a 5–10 mL sucrose gradient onto which 0.5–1 mL of cell extract is layered and centrifuged at high speed for 3–4 h in a floor-model ultracentrifuge. After centrifugation, the gradient solution is passed through an absorbance recorder to generate a polysome profile. Ten to twelve fractions (0.8–1 mL each) are collected for isolating different RNA and protein populations. The overall method is tedious and lengthy (6–9 h), requires access to a suitable ultracentrifuge rotor and centrifuge, and requires a substantial amount of tissue material, which can be a limiting factor. Moreover, there is often a dilemma over the quality of RNA and protein populations in the individual fractions due to the extended experiment times. To overcome these challenges, here we describe a miniature sucrose gradient for polysome profiling using Arabidopsis thaliana seedlings that takes ~1 h centrifugation time in a tabletop ultracentrifuge, reduced gradient synthesis time, and also less tissue material. The protocol described here can be easily adapted to a wide variety of organisms and polysome profiling of organelles, such as chloroplasts and mitochondria.
Key Features
• Mini sucrose gradient for polysome profiling that requires less than half the processing time vs. traditional methods.
• Reduced starting tissue material and sample volume for sucrose gradients.
• Feasibility of RNA and protein isolation from polysome fractions.
• Protocol can be easily modified to a wide variety of organisms (and even polysome profiling of organelles, such as chloroplast and mitochondria).
Graphical Overview
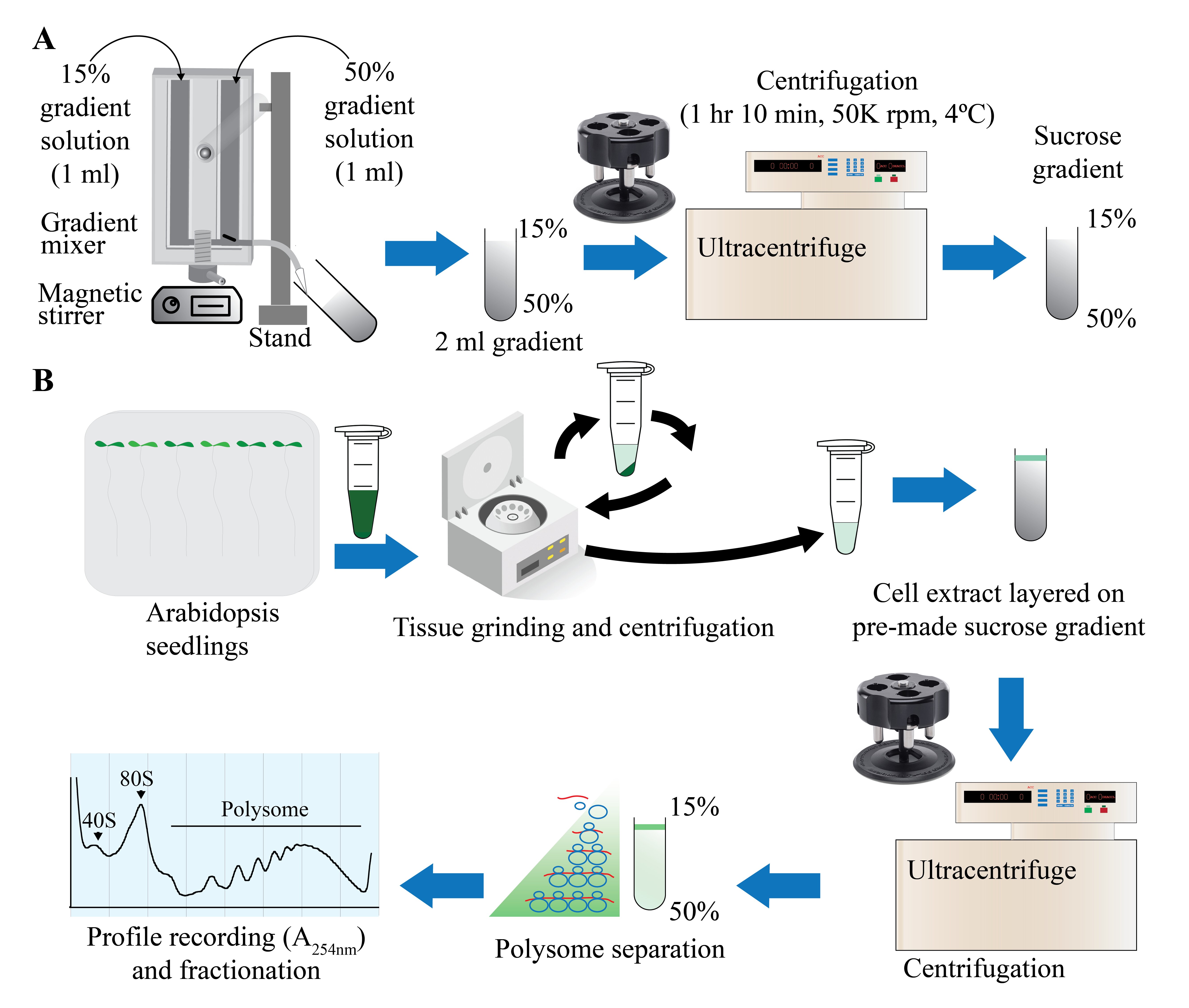
Figure 1. Graphical overview of polysome profiling using mini sucrose gradient. A. One milliliter each of 15% (w/v) and 50% (w/v) sucrose gradient solution is added to the individual chambers of the gradient maker. While mixing with a small magnetic stirrer in the 50% solution chamber, base station knob is turned to open position, allowing sucrose gradient solution to slowly flow through the outlet into a 2.2 mL gradient tube. After centrifugation at 50,000 rpm (213,626.2 × g) in a swinging bucket rotor for 70 min at 4 °C, the gradient tube is stored at 4 °C for the next steps. B. Cell extract from 12-day-old vertically grown Arabidopsis thaliana seedlings is centrifuged twice and 100 µL of supernatant is gently layered on the pre-made sucrose gradient from step A. After centrifugation as described in step A, polysome profile is obtained by feeding the gradient solution through an absorbance recorder (A254 nm). Eight (200 µL) fractions are collected for RNA and protein isolation.
Background
Gene expression control is studied at multiple levels and, among these, translational control provides unparalleled insights about the functional readout of the genome in a spatiotemporal manner (Urquidi Camacho et al., 2020). For example, translatome studies provide highly valuable information that explain differences between the transcriptome (total mRNA pool) and the proteome (changes in total protein levels). Translation is governed by the dynamic distribution of cellular mRNAs into the non-translating and actively translating pool. The actively translating mRNAs loaded with multiple 80S ribosomes (polysome) are larger in size with higher density than the non-translating mRNAs, which are either free or associated with just the 40S ribosomal subunit. Polysome profiling by sucrose gradient centrifugation is a powerful technique for studying changes related to the distribution of mRNAs, as well as genome-wide effects on the translatome (Zuccotti and Modelska, 2016).
Traditionally, for translational control studies in plants (Mustroph et al., 2009; Layat et al., 2014; Missra et al., 2015; Yamasaki et al., 2015; Bai et al., 2017; Enganti et al., 2017; Lokdarshi et al., 2020a and 2020b), the method begins with the synthesis of a continuous sucrose gradient with variable density, for example, 15%–50% (w/v) sucrose gradient solution in a 5–10 mL centrifuge tube. Around 0.5–1 g of plant tissue material is ground and 0.5–1 mL of cellular lysate in polysome extraction buffer is gently layered onto the top of the gradient. The centrifuge tubes are spun at high speed for 3–4 h at 4 °C. RNA and protein components that are smaller in size and less dense (e.g., ribosome-free mRNAs, 40S ribosome subunit) travel less far into the gradient and therefore distribute at the top of the gradient, while larger species (e.g., 60S and 80S ribosome subunit, polysomes) travel farther into the gradient. After centrifugation, the gradient tube is assembled into a fractionator and 10–12 fractions (each with 1–1.2 mL) from the top (smaller, slower traveling) to bottom (bigger, faster traveling) are collected for RNA and protein extraction. Polysome profiles are constructed by measuring the optical density of the fractions while they are fed through an absorbance recorder (254 nm). Even though the traditional procedure continues to be a gold standard for studying translational control, this protocol requires ample starting material and suffers from being too lengthy, which further raises doubts over the quality of both RNA and the protein components. To overcome some of these drawbacks, we designed and successfully showed the use of a mini sucrose gradient for polysome profiling with Arabidopsis thaliana seedlings that has tremendous advantages, such as greatly reduced gradient synthesis and centrifuge times and low sample tissue requirement (Lokdarshi et al., 2020b and 2020c). The protocol described here can be easily adapted to a wide variety of organisms (e.g., bacteria, yeast, fungus, and plant and animal tissue) and even for the polysome profiling of organelles such as chloroplasts and mitochondria.
Materials and Reagents
Biological materials
Twelve-day-old Arabidopsis thaliana ecotype Columbia (Col_0) seedlings
Solutions
DEPC-treated water
Milli-Q water treated with 0.2% (v/v) DEPC for 24 h at room temperature and autoclaved for 20 min in a glass bottle. Cool down the bottled water at room temperature for 24 h before use. (Can be stored indefinitely if the bottle is unopened)
All reagent stocks (except sucrose solution, cycloheximide, RNase inhibitor and DTT) should be made in a glass beaker and stored in a 100 mL glass bottle in the dark at room temperature. (Replace with new stock after three months)
Cycloheximide (CHX, 50 mg/mL) stock is made by resuspending 50 mg of cycloheximide in 1 mL of ethanol in a 1.5 mL tube. Store stock solution at -20 °C. (Replace with new stock after three months)
Chloramphenicol (CHL, 50 mg/mL) stock is made by resuspending 50 mg of chloramphenicol in 1 mL of ethanol in a 1.5 mL tube. Store stock solution at -20 °C. (Replace with new stock after three months)
Dithiothreitol (DTT, 1 M) stock is made by resuspending 0.154 g of DTT in 1 mL of water in a 1.5 mL tube. Store stock solution at -20 °C. (Replace with new stock after two months)
Sucrose solutions made in a glass beaker need to be filtered through a 0.2 µm PVDF filter bottle and stored in the dark at 4 °C. (Replace with new stock after one month. Before each use, always shake the bottle to check for any cloudy appearance. Discard and make new solutions as necessary)
Punch solution should be made in a glass bottle and autoclaved for 20 min on liquid cycle. (Can be stored indefinitely at room temperature in the dark)
100 mL of 1 M Tris-HCl (pH 8.4) is made by dissolving 12.14 g of Tris in 80 mL of deionized water and adjusting the pH to 8.4 using 0.1 N HCl. After attaining the desired pH, the volume of 1 M Tris-HCl solution is adjusted to final 100 mL using deionized water. (Solution is stored at room temperature and it is recommended to replace with new stock after three months)
Lysis buffer (1,000 µL in a 2 mL tube) (see Recipes)
15% (or 50%) sucrose solution (100 mL) (see Recipes)
Punch solution (100 mL) (see Recipes)
Recipes
Lysis buffer (1,000 µL in a 2 mL tube)
Make fresh from stock solutions for each experiment.
Reagent Final concentration Amount Tris-HCl (1 M, pH 8.4)
KCl (0.5 M)
MgCl2 (0.5 M)
Deoxycholic acid (10%)
Polyoxyethylene 10 tridecyl ether [20% (w/v)]
Cycloheximide (50 mg/mL)
RNase inhibitor (40 U/µL)
H2O
200 mM
50 mM
25 mM
1% (v/v)
2% (v/v)
50 µg/mL
40 U/mL
n/a
200 µL
50 µL
25 µL
100 µL
100 µL
1 µL
1 µL
523 µL
Total n/a 1,000 µL 15% (or 50%) sucrose solution (100 mL)
Reagent Final concentration Amount Tris-HCl (1 M, pH 8.4)
KCl (0.5 M)
MgCl2 (0.5 M)
Sucrose
H2O
200 mM
50 mM
25 mM
15% or 50% (w/v)
n/a
20 mL
10 mL
5 mL
15 or 50 g
40 mL
Total n/a Make up final volume to 100 mL Punch solution (100 mL)
*Note: Water should be added in increments of 10 mL. Allow the solution to mix completely before adding any more water. Sucrose will take a substantial volume when fully dissolved.
Reagent Final concentration Amount Sucrose
Bromophenol blue (0.1% (w/v))
Tris-HCl (1 M, pH 8.4)
H2O
60% (w/v)
0.02% (v/v)
1 mM
n/a
60 g
20 mL
100 µL
see note*
Total n/a 100 mL
Laboratory Supplies
1.5 and 2 mL tubes (Thermo Fisher, catalog numbers: AM12450 and AM12475)
Pipettes (200–1,000, 20–200, 2–20, and 0.2–2 µL)
DNase/RNase free tips (1,000, 200, 10 µL)
1.5 mL tube rack
7/16 × 13/8 in. (11 × 34 mm) 2.2 mL polypropylene tube (Beckman Coulter, catalog number: 347357)
19 Gauge needle for gradient tube piercing (Fisher Scientific, catalog number: 14-826-52)
Parafilm (Fisher Scientific, catalog number: S37440)
Disposable bottle top filters (Thermo Fisher, catalog number: 597-4520)
Disposable bottle filter units (Thermo Fisher, catalog number: 568-0020)
Permanent ink markers
Clear tubing (6 inch in length) for gradient mixer outlet (1/16 in diameter)
Wipes (Fisher Scientific, catalog number: 06-666C)
Flat-tip forceps (Fisher Scientific, catalog number: 16-100-116)
Porcelain pestle and mortar (Fisher Scientific, catalog number: S27075)
Ice bucket (Styrofoam box can be used as an alternative)
Basic binder paper clip (medium)
Gloves
Stainless steel spatula with tapered end
Tris (Molecular Grade) (Promega, Fisher Scientific catalog number: PR-H5133)
KCl (Fisher Scientific, catalog number: AC418205000)
MgCl2 (Fisher Scientific, catalog number: M33-500)
Deoxycholic acid (Fisher Scientific, catalog number: BP349-100)
Polyoxyethylene 10 tridecyl ether (Millipore Sigma, catalog number: P2393-100G)
Cycloheximide (Millipore Sigma, catalog number:01810-G)
Chloramphenicol (Millipore Sigma, catalog number: 220551-25GM)
40 U/mL RNase inhibitor (Promega, catalog number: N2515)
Sucrose, molecular biology grade (Millipore Sigma, catalog number: 573113)
Bromophenol blue (Fisher Scientific, catalog number: AAA1846909)
Dithiothreitol (DTT) (Thermo Fisher Scientific, catalog number: R0861)
Murashige and Skoog (MS) plant basal salt (MP Biomedicals, Fisher Scientific catalog number: ICN2623022)
RNaseZapTM RNase decontamination solution (ThermoFisher, catalog number: AM9780)
Liquid nitrogen
Aluminum foil
Equipment
Gradient mixer (Buchler Instruments). Catalog number for this specific product is not available as no vendor supports this product anymore. For custom manufacturing of the gradient mixer, see Figure 2.
Tabletop ultracentrifuge (Beckman Coulter, catalog number: A95761 or equivalent)
TLS-55 swinging-bucket rotor with four buckets for up to four gradients (Beckman Coulter, catalog number: 346936)
Gradient station or any equivalent gradient fractionators (Biocomp, catalog number: 153-002)
ISCO UA 5 100 absorbance/fluorescence monitor. Catalog number for this specific product is not available as the vendor is out of market. Alternative product information: BR-188 Density Gradient Fractionation System with Peak Chart Data Acquisition System (Brandel)
Tabletop centrifuge with rotor to hold 1.5 mL tube. If refrigerated centrifuge is not available, then a non-refrigerated centrifuge can be placed in a 4 °C cold room as an alternative
Magnetic plate stirrer
Magnetic stirrer bar (Fisher Scientific, catalog number: 14-513-93). Cut the magnetic stirrer bar into half with the plier and store the halves in a 1.5 mL tube (micro-stirrer bar).
Note: Cleaning of this micro stirrer bar should be done with warm water and stored carefully.
Weighing balance with 0.001 g accuracy
Vortex
Burette stand
Software and Datasets
The BR-188 Density Gradient Fractionation System comes with the Peak Chart Data Acquisition System Software. See note in section E to develop polysome profile without a Peak Chart Data Acquisition System Software.
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Lokdarshi, A. and von Arnim, A. G. (2023). A Miniature Sucrose Gradient for Polysome Profiling. Bio-protocol 13(6): e4622. DOI: 10.21769/BioProtoc.4622.
- Lokdarshi, A., Papdi, C., Pettko-Szandtner, A., Dorokhov, S., Scheres, B., Magyar, Z., von Arnim, A. G., Bogre, L. and Horváth, B. (2020c). ErbB-3 BINDING PROTEIN 1 Regulates Translation and Counteracts RETINOBLASTOMA RELATED to Maintain the Root Meristem. Plant Physiol 182(2): 919-932.
分类
植物科学 > 植物分子生物学 > 遗传分析
分子生物学 > RNA > mRNA 转译
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link