- EN - English
- CN - 中文
Molecular Surveillance of Malaria Using the PF AmpliSeq Custom Assay for Plasmodium falciparum Parasites from Dried Blood Spot DNA Isolates from Peru
使用 PF AmpliSeq定制检测法对秘鲁干血斑 DNA 中的恶性疟原虫寄生虫分子进行疟疾监测
发布: 2023年03月05日第13卷第5期 DOI: 10.21769/BioProtoc.4621 浏览次数: 2117
评审: Alka MehraMariana Barnes Rita Marie Celine Meganck
Abstract
Malaria molecular surveillance has great potential to support national malaria control programs (NMCPs), informing policy for its control and elimination. Here, we present a new three-day workflow for targeted resequencing of markers in 13 resistance-associated genes, histidine rich protein 2 and 3 (hrp2&3), a country (Peru)-specific 28 SNP-barcode for population genetic analysis, and apical membrane antigen 1 (ama1), using Illumina short-read sequencing technology. The assay applies a multiplex PCR approach to amplify all genomic regions of interest in a rapid and easily standardizable procedure and allows simultaneous amplification of a high number of targets at once, therefore having great potential for implementation into routine surveillance practice by NMCPs. The assay can be performed on routinely collected filter paper blood spots and can be easily adapted to different regions to investigate either regional trends or in-country epidemiological changes.
Keywords: Plasmodium falciparum (恶性疟原虫)Background
Surveillance of Plasmodium parasites has been identified by the World Health Organization (WHO) as one of the essential pillars to move towards malaria elimination in its endemic areas (WHO, 2019). Molecular parasite genotyping tools can strengthen malaria surveillance systems by monitoring the emergence and spread of drug resistance, histidine rich protein 2 and 3 (hrp2 and hrp3) deletions, quantification of malaria importation risk, and characterization of changing transmission intensity. As molecular surveillance platforms transition from proof-of-concept research studies to operational incorporation into national malaria control programs (NMCPs), simple standardized laboratory protocols feasible on benchtop sequencers and automated analysis pipelines are necessary to generate reproducible results and decrease the time from sample collection to results, thereby ensuring rapid turnover of up-to-date reports for decision making and policy (WHO, 2016; MPA Committee, 2019). Various methods have been developed to investigate drug resistance markers, usually based on the amplification of loci of interest using PCR-based techniques, with amplicons detected in real time assays by amplicon sequencing (Taylor et al., 2013; Menard et al., 2016; Nsanzabana et al., 2018), or analyzed using restriction fragment length polymorphism (Eldin de Pecoulas et al., 1995; Duraisingh et al., 1998). In the past, population surveillance widely utilized genotyping tools targeting surface antigens by PCR to distinguish parasite clones (Falk et al., 2006; Koepfli et al., 2009), followed by panels of microsatellites (MS) that are not under evolutionary selection pressure and are, therefore, more suitable to inform population genetic changes (Anderson et al., 2000; Imwong et al., 2007; Karunaweera et al., 2008). More recently, genome-wide single nucleotide polymorphism (SNP) panels, capable of defining a molecular barcode to capture the diversity of parasite populations, have been developed and can be investigated with methods such as microarrays, real-time PCR, and deep sequencing (Daniels et al., 2008, 2013 and 2015; Neafsey et al., 2008; Baniecki et al., 2015; Koepfli and Mueller, 2017; Fola et al., 2020; Tessema et al., 2022). Surveys to detect these gene deletions rely on molecular methods based on PCR assays that classify deletions based on failure to amplify targets in the hrp2 and hrp3 exon region (and sometimes flanking genes).
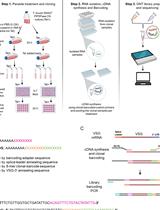
Currently, there is no multifunctional tool that includes a combination of more than two types of markers (i.e., SNP-barcodes, drug resistance, etc.) to serve several use cases. In addition, the characterization of hrp2 and hrp3-deletions relies on PCR assays that classify deletions based on failure to amplify targets. The few existing tools that combine population markers with drug resistance target short regions around validated drug resistance SNPs, missing the potential to detect novel resistance-associated mutations. Furthermore, many SNP-panels were designed from genomes across the world and lack the resolution to study subtle patterns on a smaller geographical scale. Therefore, we have developed a targeted amplicon next-generation sequencing (NGS) assay for molecular surveillance of Plasmodium falciparum parasites that combines a specifically designed barcode for the target country, 13 full-length resistance-associated genes (Table 1), hrp2 and hrp3, and an apical membrane antigen 1 (ama1) microhaplotype region. The novelty of the assay is its high number of targets multiplexed in one easy workflow, using AmpliSeq deep sequencing technology (Illumina, 2018), and combining phenotypic markers with a 28-SNP barcode with in-country resolution to investigate parasite gene flow in Peru. The assay uses overlapping amplicons to cover large genes. The 28 SNP-barcode was designed with in-country resolution to monitor parasite strains circulating in Peru over space and time (Kattenberg et al., 2023). These SNPs were selected from a South American P. falciparum genome dataset selecting SNPs with a frequency of minor alleles below 0.35 and those that were not under selective pressure, prioritizing SNPs that differentiated the Peruvian samples from other parasite populations in the dataset. Noteworthy, the seq-based tool can be performed on routinely collected filter paper blood spots and can be easily adapted to different regions to investigate either regional trends or in-country epidemiological changes using different SNP-barcodes. The technology applies a multiplex PCR approach to amplify all genomic regions of interest in a rapid and easily standardizable procedure, and allows simultaneous amplification of a high number of targets at once, therefore having great potential for implementation into routine surveillance practice by NMCPs.
Table 1. Targeted genes of interest for drug resistance in the PF AmpliSeq assay
| Gene ID | Chromosome | Gene with resistance | Drug associated with resistance* |
| PF3D7_1218300 | 12 | ap2-mu | ART; QN |
| PF3D7_1251200 | 12 | coronin | DHA |
| PF3D7_0709000 | 7 | crt | CQ; PPQ; AMQ |
| mal_mito_3 | mitochondrial | cytochrome B | ATQ |
| PF3D7_0417200 | 4 | dhfr | PYR; PG |
| PF3D7_0810800 | 8 | dhps | SULF |
| PF3D7_1362500 | 13 | exonuclease | PPQ |
| PF3D7_1343700 | 13 | k13 | ART |
| PF3D7_0523000 | 5 | mdr1 | CQ; PPQ; MQ; QN; HF; AMQ; LF |
| PF3D7_0112200 | 1 | mrp1 | ART, MQ, LF |
| PF3D7_1408000 | 14 | plasmepsin II | PPQ |
| PF3D7_API04900 | apicoplast | 23s rRNA | CM |
| PF3D7_0104300 | 1 | ubp-1 | ART |
AMQ: amodiaquine; ART: artesunate; ATQ: atovaquone; CM: clindamycin; CQ: chloroquine; DHA: dihydroartemisinin; HF: halofantrine; LF: lumefantrine; MQ: mefloquine; PG: proguanil; PPQ: piperaquine; PYR: pyrimethamine; SULF: sulfadoxine; QN: quinine. *The list of drugs to which resistance has been observed is non-exhaustive. Validated mutations are listed in the WHO report on antimalarial drug efficacy, resistance, and response: 10 years of surveillance (2010–2019).
Materials and Reagents
AmpliSeq Library PLUS kit for Illumina (96 reactions; Illumina, catalog number: 20019102)
Contains:
1× Lib Amp mix
10× Library Amp primers
DNA ligase
5× AmpliSeq HiFi mix
FuPa Reagent
Low TE (Tris-EDTA buffer)
Switch solutions
AmpliSeq Custom DNA panel for Illumina (Illumina, catalog number: 20020495, custom design IAD179763_241; see Supplementary manifest file and APPENDIX 1 for oligo sequences)
Contains:
Primer pool 1 (red cap)
Primer pool 2 (blue cap)
AmpliSeq CD Indexes Set A for Illumina (96 indexes, 96 samples; Illumina, catalog number: 20019105)
Note: Optionally, you can also use Index Set B, C, or D.
Absolute ethanol (EtOH) (Sigma-Aldrich, Merck, catalog number: 1.00983.1000)
Agencourt AMPure XP beads (Beckman Coulter, catalog number: A63881)
96-well PCR plate, 0.2 mL (Greiner Bio-One, catalog number: 652201)
8-well PCR strips (Greiner Bio-One, catalog number: 673210)
Strip caps for PCR pates (Greiner Bio-One, catalog number: 373250)
Adhesive seals for PCR plates (Westburg Life Science, catalog number: WB2-3800)
1.5 mL DNA LoBind® Tubes (Eppendorf, catalog number: 022431021)
Nuclease-free water (Lonza, catalog number: BE51200)
KAPA Library Quantification kit (KAPA Biosystems, Roche, catalog number: 07960298001)
DNA standards 1–6 (80 µL each)
Primer mix (1 mL)
KAPA SYBR® FAST qPCR Master mix (5 mL)
Qubit® dsDNA HS Assay kits (Invitrogen, Thermo Fischer Scientific, catalog number: Q32851)
QubitTM dsDNA HS reagent (250 µL)
QubitTM dsDNA HS buffer (50 mL)
QubitTM dsDNA HS standard #1 (1 mL)
QubitTM dsDNA HS standard #2 (1 mL)
Thin-wall, clear, 0.5 mL PCR tubes (e.g., Qubit® assay tubes, catalog number: Q32856)
Pipette filter tips, 10 µL (e.g., Greiner Bio-One, catalog number: 771353; Biofil, catalog number: PPT150010)
Combitips 0.2 and 0.5 mL (Eppendorf, catalog numbers: 0030089774 and 0030089421)
Miseq Reagent kit v3 (600 cycle) (Illumina, catalog number: MS-102-3003)
Reagent cartridge
HT1 (hybridization buffer, 5 mL)
PR2 (incorporation buffer, 500 mL)
Flow cell
1 N sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: 22146-5)
Tris-hydrochloride (HCl), pH 8.0(Sigma-Aldrich, catalog number: T3038-1L) , adjusted to pH 7.0
Tween® 20 (Sigma-Aldrich, catalog number: 022431021)
PhiX Control v3 (Illumina, catalog number: FC-110-3001)
Optional to include as controls and take along in the library preparation:
DNA extracted from laboratory strain(s) of P. falciparum (e.g., 3D7 or Dd2) as positive control
Note: Laboratory strains can be obtained from BEI Resources, NIAID, NIH (https://www.beiresources.org/): P. falciparum, strain 3D7, MRA-102, contributed by Daniel J. Carucci; P. falciparum, strain Dd2, MRA-150, contributed by David Walliker; P. falciparum, strain Dd2_R539T, MRA-1255 and P. falciparum, strain CamWT_C580Y, MRA-1251, contributed by David A. Fidock; and P. falciparum, strain IPC 4912, MRA-1241, contributed by Didier Ménard. Genomic DNA of these strains is also available from BEI Resources.
DNA extracted from uninfected human blood as negative control
Equipment
Multichannel pipettes 10 µL, 0.1 mL and 0.3 mL
Electronic multi-dispenser pipette (Eppendorf, Multipette © E3x, catalog number: 4987000029)
Centrifuge (Sigma Laborzentrifugen GmbH, Sigma 4-16KS, catalog number: 150507)
Vortex (Scientific Industries, Vortex Genie 2, catalog number: SI-0236)
96-well magnet stand (Invitrogen, ThermoFisher Scientific, DynamagTM-96 Side Skirted, #12027; RNA Life Technologies, Ambion® Magnetic Stand-96, catalog number: 1307065)
Qubit 2.0 (or higher) fluorometer (Invitrogen, Thermo Fischer Scientific, catalog number: Q32866)
Conventional cycler (Biometra, Westburg, Professional Thermocycler Basic 96 × 0.2 mL, catalog number: 846-070-701)
LightCycler® 480 real-time PCR system (Roche, catalog number: 04640268001)
MiSeqTM system (Illumina, catalog number: SY-410-1003)
Software
Illumina experiment Manager (Illumina)
Microsoft Excel (Microsoft)
In-house Linux-based pipeline
Ubuntu (Canonical, ubuntu.com)
Trimmomatic (Bolger et al., 2014, https://doi.org/10.1093/bioinformatics/btu170)
Burrows Wheeler Aligner (Li et al., 2009, https://doi.org/10.1093/bioinformatics/btp324)
Picard tools (Broad Institute, https://broadinstitute.github.io/picard/)
Genome Analysis Toolkit (GATK) (Broad Institute, https://gatk.broadinstitute.org/)
SnpEff (Cingolani et al., 2012)
On-Off instrument alignment
Local Run Manager (Illumina)
DNA Amplicon Module (Illumina)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Kattenberg, J. H., van Dijk, N. J., Fernández-Miñope, C. A., Guetens, P., Mutsaers, M., Gamboa, D. and Rosanas-Urgell, A. (2023). Molecular Surveillance of Malaria Using the PF AmpliSeq Custom Assay for Plasmodium falciparum Parasites from Dried Blood Spot DNA Isolates from Peru. Bio-protocol 13(5): e4621. DOI: 10.21769/BioProtoc.4621.
分类
分子生物学 > DNA > DNA 测序
微生物学 > 微生物遗传学 > DNA
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link