- EN - English
- CN - 中文
A Novel Method to Isolate RNase MRP Using RNA Streptavidin Aptamer Tags
使用 RNA 链霉亲和素适体标签分离 RNase MRP 的新方法
(*contributed equally to this work) 发布: 2023年02月20日第13卷第4期 DOI: 10.21769/BioProtoc.4615 浏览次数: 2919
评审: Alessandro DidonnaKirsten A. CoprenAnonymous reviewer(s)

相关实验方案

适用于 LC–MS/MS 蛋白质组学分析的大体积细胞培养上清分泌组样品制备方法优化
Basil Baby Mattamana [...] Peter Allen Faull
2025年12月20日 1260 阅读
Abstract
Interactions between RNA-binding proteins and RNA molecules are at the center of multiple biological processes. Therefore, accurate characterization of the composition of ribonucleoprotein complexes (RNPs) is crucial. Ribonuclease (RNase) for mitochondrial RNA processing (MRP) and RNase P are highly similar RNPs that play distinct roles at the cellular level; as a consequence, the specific isolation of either of these complexes is essential to study their biochemical function. Since their protein components are nearly identical, purification of these endoribonucleases using protein-centric methods is not feasible. Here, we describe a procedure employing an optimized high-affinity streptavidin-binding RNA aptamer, termed S1m, to purify RNase MRP free of RNase P. This report details all steps from the RNA tagging to the characterization of the purified material. We show that using the S1m tag allows efficient isolation of active RNase MRP.
Keywords: Ribonucleoprotein (核糖核蛋白)Background
In order to maintain RNA functions or defend the organism, most RNAs can be the substrate of ribonucleases (RNases), enzymes specialized in RNA cleavage and degradation. Ribonucleases are generally composed of a catalytic RNA-binding protein, but the catalyst can also be an associated RNA molecule, which is called a ribozyme. In the latter, the catalytic RNA is generally associated with one or more proteins, together forming a ribonucleoprotein particle (RNP). Two RNP-type ribonucleases in humans are RNase for mitochondrial RNA processing (MRP) and RNase P (Guerrier-Takada et al., 1983), complexes with a highly similar structure. These endoribonucleases, essential to cell survival, are ubiquitously expressed RNPs and are mostly located in the nucleolus of eukaryotic cells. RNase MRP and RNase P are evolutionarily related; RNase P is present in all domains of life, while RNase MRP is only found in eukaryotes (Randau et al., 2008).
RNase MRP and RNase P both contain a unique long non-coding RNA, but share most of their protein subunits. Ten protein subunits (hPop1, hPop4, hPop5, Rpp14, Rpp20, Rpp21, Rpp25, Rpp30, Rpp38, and Rpp40) have been demonstrated to interact with both particles, although hPop4, Rpp14, and Rpp21 might be more strongly associated with RNase P (Welting et al., 2006). The RNA components of RNase MRP and RNase P differ in size and sequence, but they share the same conserved domains and fold into highly similar secondary structures. Despite their similarity, RNase MRP and RNase P have very distinct functions. While RNase MRP, named after its first discovered role in Mitochondrial RNA Processing (Chang and Clayton, 1987), is also involved in ribosomal RNA (rRNA) (Lygerou et al., 1996; Lan et al., 2020) and messenger RNA (mRNA) maturation (Gill et al., 2004; Mattijssen et al., 2011), RNase P is best known for its function as a processor of transfer RNA (tRNA) precursors (Robertson et al., 1972). In addition to the maturation of pre-tRNA, RNase P has been reported to be involved in transcription by RNA polymerase I and III (Reiner et al., 2006; 2008), as well as in the maturation of small nucleolar RNAs (snoRNAs) in yeast (Coughlin et al., 2008).
Mutations in the RNase MRP RNA (RMRP) cause a spectrum of disorders characterized by similar symptoms, the major condition being the autosomal recessive disease cartilage-hair hypoplasia (CHH, OMIM #250250). This pathology is mostly characterized by severe skeletal dysplasia and multiple pleiotropic symptoms (Hermanns et al., 2006; Mattijssen et al., 2010). CHH-causing mutations, mainly single-nucleotide substitutions, are hypothesized to affect RMRP function by altering its secondary structure or its interaction with other molecules (Welting et al., 2008). Today, the molecular mechanisms of CHH pathology remain elusive. In order to better comprehend the cellular function of RNase MRP and the disorders linked to mutations in its RNA subunit, it is necessary to gain knowledge about the composition of this RNP and its interactome.
A ribonucleoprotein complex is commonly purified by either a protein-centric or an RNA-centric method, depending on which molecule is being targeted (Ramanathan et al., 2019). Protein-centric approaches, such as immunoprecipitation or the use of protein affinity tags, allow specific purification of some major RNA–protein cellular complexes. However, this is only possible if they contain a unique protein component, e.g., a protein subunit absent from any other RNP complex. No protein component unique to the human RNase MRP has been discovered to date and, as a consequence, any protein-centric method employed to purify the RNase MRP complex will also co-purify RNase P. This will result in a mixture of both enzymes in the purified samples, and the inability to differentiate between the two RNPs leads to uncertainties on their biochemical function. For instance, it was demonstrated that RNase MRP or RNase P is responsible for the endoribonucleolytic cleavage of m6A-modified RNAs (Park et al., 2019), but differentiation between these two endoribonucleases was not possible, since the purification was based on a shared protein subunit.
When RNP complexes cannot be isolated by techniques targeting their protein components with sufficient specificity, RNA-centric methods can be employed. However, the non-coding RNA subunits of both RNase MRP and RNase P are relatively short, highly structured, and shielded with proteins. This makes it quite challenging to purify them with RNA-specific probes, such as biotin-labeled antisense oligonucleotides (Hou et al., 2016).
As an alternative to antisense oligonucleotides, an RNA aptamer can be inserted in the RNA molecule of interest to serve as a tag for purification. Several RNA aptamers that bind ligands with high affinity have been developed in the last decades, making them powerful tools for RNA-specific purification (Walker et al., 2008). Examples include the StrepTag (Bachler et al., 1999), the MS2-TRAP (MS2-tagged RNA affinity purification) procedure (Yoon et al., 2012), and streptavidin aptamer (S1) tagging. Insertion of MS2 hairpins has been proven useful for the characterization of complexes containing a non-coding or messenger RNA of interest, but it requires co-expression of a tagged MS2-binding protein (Yoon et al., 2012; Yoon and Gorospe, 2016).
The S1 aptamer was identified by in vitro selection as a streptavidin-binding short sequence with a high affinity and appeared to be effectively eluted with biotin for the successful recovery of the tagged molecule (Srisawat and Engelke, 2001). Widely available streptavidin-agarose beads can be used, and this tag has been shown to allow efficient purification of RNase P from yeast (Srisawat and Engelke, 2002) and human cells (Li and Altman, 2002).
In this report, we describe a novel method to specifically isolate RNase MRP using the S1m RNA aptamer (Derksen et al., 2022), an optimized version of the original S1 aptamer (Leppek and Stoecklin, 2014). In addition to the increase in purification efficiency compared to the S1 tag, the S1m aptamer has a relatively short size. This lowers the risk of undesirable effects on the secondary and tertiary structure of the RNA of interest, known to be crucial for the function of many non-coding RNAs, including ribozymes.
Briefly, the S1m aptamer is inserted in the RNA of interest by cloning and the purified plasmid is used to transfect human cells. The complex containing the tagged RNA is purified from the lysate using streptavidin-coated beads and all proteins and RNAs are extracted from the purified material. The composition of the bound material is then identified by northern and western blotting methods (Figure 1).
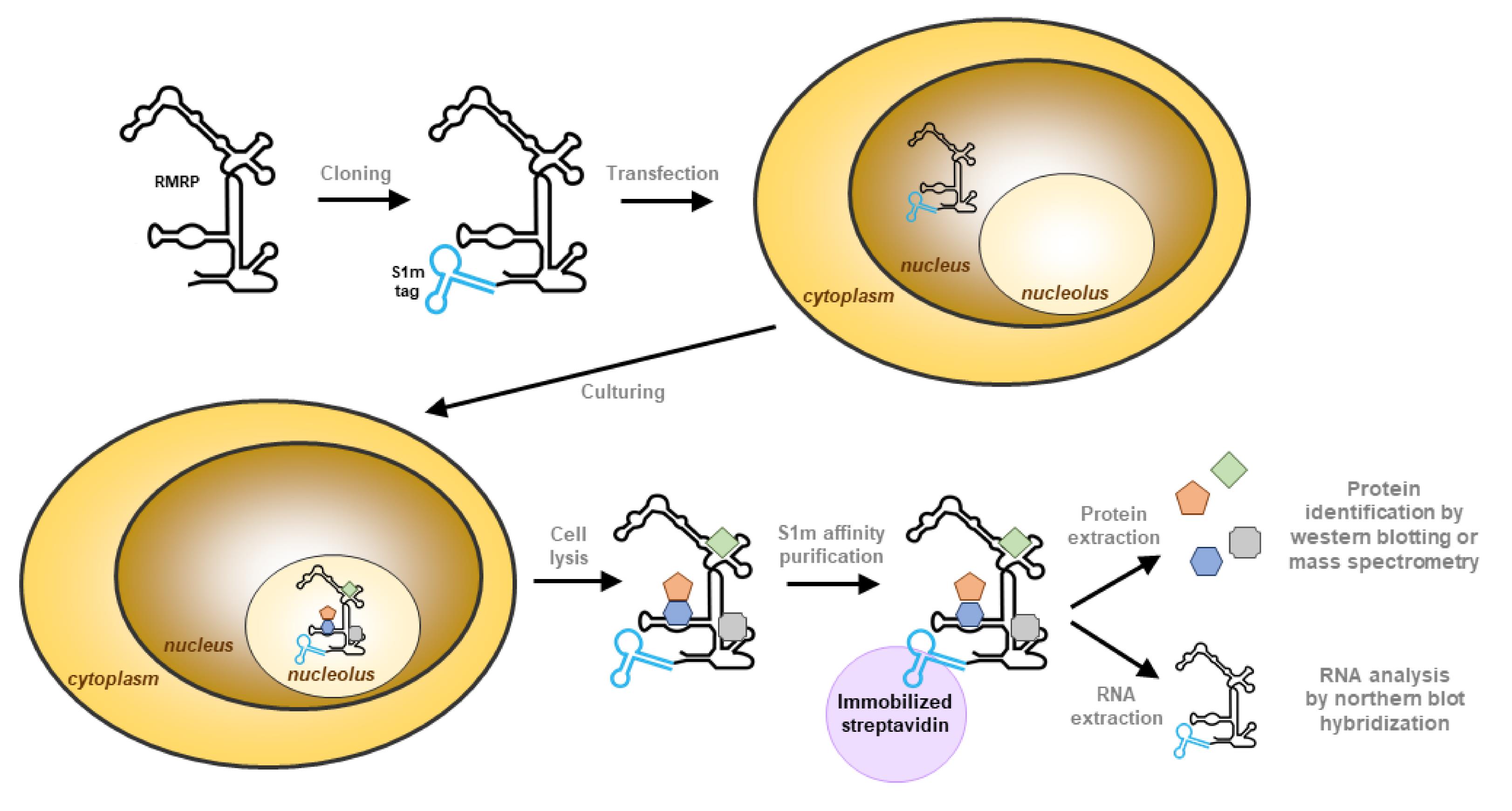
Figure 1. Schematic illustration of the isolation of RNase MRP using the S1m tag. The S1m aptamer is introduced at the desired location of the gene of interest (5′, 3′, or internal) and human cells are transfected with this construct. The tagged RNA is transcribed in the nucleus and will assemble into ribonucleoprotein particles, leading to accumulation in the nucleoli. The cells expressing the tagged RNA are lysed, and the RNA–protein complex is purified with immobilized streptavidin, which binds to the S1m tag. The purified complex is eluted from the beads to extract the proteins and RNA separately. The RNA sample can be used to analyze the specificity of purification by northern blot hybridization and the protein sample can be used to analyze the associated proteins.
Materials and Reagents
Cell lines
Human embryonic kidney HEK293T cells
Disposables
For all RNA work, use only filter tips, RNA- and RNase-free tubes, and prepare all solutions RNase-free.
1.5 mL microcentrifuge tubes (autoclaved) (any vendor)
0.2 mL PCR tubes (any vendor)
Sterile 15 mL polypropylene centrifuge tubes (any vendor)
Sterile 50 mL polypropylene centrifuge tubes (any vendor)
Sterile nuclease-free filter tips (10 µL, 200 µL, 1,000 µL) (any vendor)
Sephadex G50 column (Cytiva, catalog number: 27534001)
Cell culture 12-well plate (Greiner Bio-one, catalog number: 665180)
Cell culture dishes, 150 cm2 (Greiner, catalog number: 639160)
Serological pipette, 25 mL (any vendor)
Serological pipette, 10 mL (any vendor)
Serological pipette, 5 mL (any vendor)
Petri dishes (Sarstedt, catalog number: 82.1473.001)
Sterile surgical blades for the excision of DNA bands from gel (Swann-Morton, catalog number: 0208)
QIAquick gel extraction kit protocol (Qiagen, catalog number: 28706)
Plasmid Midi kit (Qiagen, catalog number: 12145)
Hybond-N membrane (GE Healthcare, catalog number: RPN303B)
Protran BA 85 nitrocellulose membrane, 0.45 µm pore size (VWR, catalog number: 10063-173)
Filter paper (any vendor)
Whatman paper 0.34 mm (Cytiva, catalog number: 3030-917)
0.22 µm filter (VWR, catalog number: 514-0061)
Reagents (order of use)
Sterile double distilled water (DDW), referred to as ultrapure or RNase-free water
Tryptone (MP Biomedicals, catalog number: 091010817)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014)
Yeast extract (MP Biomedicals, catalog number: 0210330391)
Agar (Thermo Scientific, catalog number: 30391023)
Sodium hydroxide (NaOH) EMSURE® (VWR, catalog number: 1.06498.1000)
Agarose (Eurogentec, catalog number: EP-0010-05)
DNA loading dye, 6× (Thermo Scientific, catalog number: R0611)
Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Scientific, catalog number: 41966)
Penicillin-streptomycin solution (Thermo Scientific, catalog number: 151401)
Fetal calf serum (FCS) (Sigma-Aldrich, catalog number: F2442)
Polyethyleneimine (PEI), branched (Sigma-Aldrich, catalog number: 108727)
Opti-minimal essential medium (Opti-MEM) (Gibco, catalog number: 51985026)
Potassium chloride (KCl) (Thermo Fisher Scientific, catalog number: 15496249)
Sodium phosphate monobasic monohydrate (NaH2PO4·H2O) (VWR, catalog number: MERC1.06346)
Sodium hydrogen carbonate (NaHCO3) EMSURE® (VWR, catalog number: 1.06329)
Glucose (Invitrogen, catalog number: 15023021)
Trypsin (Sigma-Aldrich, catalog number: T4799)
Ethylenediaminetetraacetic acid (EDTA) (VWR, catalog number: 1.08418)
Dulbecco’s phosphate buffered saline (PBS) (DPBS) (Life Technologies, catalog number: 14190)
Tris (Sigma-Aldrich, catalog number: T1378)
Hydrochloric acid (HCl), 37% (VWR, catalog number: 1.00317)
Glycerol (Bio-Connect, catalog number: 4800688)
NP-40 (IGEPAL®) (Sigma-Aldrich, catalog number: 56741)
cOmplete protease inhibitor cocktail (Roche, catalog number: 1697498001)
RNasin (Promega, catalog number: N2111)
Streptavidin-conjugated SepharoseTM beads (GE Healthcare, catalog number: 17511301)
Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L4390)
β-Mercaptoethanol (β-ME) (Sigma-Aldrich, catalog number: M3148)
Bromophenol blue (BFB) (Sigma-Aldrich, catalog number: B5525)
Trizol reagent (Invitrogen, catalog number: 15596018)
Chloroform, EMSURE® (Sigma-Aldrich, catalog number: 1.02445)
Isopropanol, EMSURE® (Sigma-Aldrich, catalog number: 1.09634)
GlycoBlue (Thermo Scientific, catalog number: AM9515)
25:24:1 phenol:chloroform:isoamyl alcohol (Invitrogen, catalog number: 15593049)
Ethanol absolute, EMSURE® (Sigma-Aldrich, catalog number: 1.00986)
Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D9779)
5× transcription optimized buffer (Promega, catalog number: P1181)
100 mM NTPs (Affymetrix, catalog number: 77245)
T7 RNA polymerase (Thermo Fisher Scientific, catalog number: EP0111)
32P-α-UTP (PerkinElmer, 3000 Ci/mmol, NEG502Z)
Boric acid (Boom, catalog number: 61000501.1000)
40% (w/v) acrylamide:bisacrylamide (19:1) solution for RNA (Serva, catalog number: 10.679.02)
Urea (Invitrogen, catalog number: 15505027)
N,N,N’,N’-Tetramethylethylenediamine (TEMED) (Sigma-Aldrich, catalog number: T7024)
Ammonium persulfate (APS) (VWR, catalog number: 97064-594)
Xylene cyanol FF (XCFF) (Sigma-Aldrich, catalog number: X4126)
Monosodium phosphate (NaH2PO4) (Sigma-Aldrich, catalog number: L2887)
Disodium hydrogen phosphate (Na2HPO4) (VWR, catalog number: 1.06580.1000)
Sodium citrate (VWR, catalog number: 1.06448)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A4503)
Ficoll® 400 (VWR, catalog number: 17-0300)
Polyvinylpyrrolidone (Sigma-Aldrich, catalog number: PVP40)
Herring sperm DNA (Sigma-Aldrich, catalog number: D6898)
30% (w/v) acrylamide:bisacrylamide (37.5:1) solution (Serva, catalog number: 10.688.02)
Glycine (Merck, catalog number: 50046)
Methanol (Thermo Scientific, catalog number: 11976961)
Ponceau-S solution (Sigma-Aldrich, catalog number: P7170)
Tween-20 (Merck, catalog number: 822184)
Non-fat dry milk (any vendor)
IRDye-conjugated secondary antibodies (LiCOR Biosciences)
Luria-Bertani (LB) medium and agar (see Recipes)
Complete DMEM medium (see Recipes)
10× tyrode solution (see Recipes)
Trypsin-EDTA solution (see Recipes)
Polyethyleneimine (PEI) (see Recipes)
S1m lysis buffer (see Recipes)
S1m incubation buffer (see Recipes)
S1m wash buffer (see Recipes)
4× protein sample buffer (see Recipes)
In vitro transcription (IVT) NTP mix (see Recipes)
1× TBE (see Recipes)
RNA denaturing gel (see Recipes)
2× RNA sample buffer (see Recipes)
Blotting buffer (see Recipes)
20× SSC (see Recipes)
100× Denhardt’s (see Recipes)
10 mg/mL sheared herring sperm DNA (see Recipes)
Pre-hybridization buffer (see Recipes)
SDS-PAGE running gel (see Recipes)
SDS-PAGE stacking gel (see Recipes)
SDS-PAGE running buffer (see Recipes)
Western blotting buffer (see Recipes)
Blot blocking solution (see Recipes)
PBST (see Recipes)
Optional: BioLux® Gaussia Luciferase assay kit (New England BioLabs, catalog number: E3300)
Optional: Luminometer-compatible 96-well plate, opaque white or black (any vendor)
Plasmids
pGEM-3Zf(+)-RMRP (Pluk et al., 1999; available by request to the authors)
pcDNA5/FRT/TO (Thermo Fisher, catalog number: V652020)
Bacterial strains
E. coli TOP10 competent cells (homemade)
Equipment
Benchtop centrifuge (Eppendorf, catalog number: 5415D)
Refrigerated benchtop centrifuge (Eppendorf, catalog number: 5417R)
Refrigerated centrifuge suitable for 50 mL volume (Heraeus Megafuge 1.0 R)
Magnetic stirrer (IKA REO Basic C IKAMAG)
Thermocycler (Bio-Rad T100)
Shaker incubator (Infors HT Multitron 2)
Spectrophotometer (DeNovix DS-11)
Gel scanner (FLA-5100)
Humidified 37 °C, 5% CO2 incubator (Heracell 150)
Diagenode bioruptor
Digital rotary mixer (Labinco, LD-76)
Standard equipment for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
Odyssey imaging system (LiCOR Biosciences)
Crosslinker, Stratalinker 1800 (Stratagene) with UV bulbs, λ = 254 nm (Ushio, catalog number: 3000016)
Geiger counter (Thermo Scientific, Mini monitor 900)
Phosphor imaging screens (Bio-Rad)
Typhoon imager (GE Healthcare)
Optional: Luminometer (Thermo Scientific, VL0000D0)
Procedure
文章信息
版权信息
© 2023 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Charteau, V., Derksen, M. and Pruijn, G. J. (2023). A Novel Method to Isolate RNase MRP Using RNA Streptavidin Aptamer Tags. Bio-protocol 13(4): e4615. DOI: 10.21769/BioProtoc.4615.
分类
生物化学 > 蛋白质 > 分离和纯化
分子生物学 > RNA > RNA 检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











