- EN - English
- CN - 中文
Preparation of Caenorhabditis elegans for Scoring of Muscle-derived Exophers
用于肌肉源性外显体评估的秀丽隐杆线虫的制备
发布: 2023年01月05日第13卷第1期 DOI: 10.21769/BioProtoc.4586 浏览次数: 1877
评审: Andrea PuharRama Reddy GoluguriAnonymous reviewer(s)

相关实验方案

用Argonaute NRDE-3标记活性转录位点—图像秀丽隐杆线虫体内的活性转录位点
Antoine Barrière and Vincent Bertrand
2022年06月05日 2686 阅读
Abstract
Utilizingresources available from the mother's body to guarantee healthy offspring growth is the fundamental reproductive strategy. Recently, we showed that a class of the largest extracellular vesicles known as exophers, which are responsible for the removal of neurotoxic components from neurons (Melentijevic et al., 2017) and damaged mitochondria from cardiomyocytes (Nicolás-Ávila et al., 2020), are released by the Caenorhabditis elegans hermaphrodite body wall muscles (BWM), to support embryonic growth (Turek et al., 2021). Employing worms expressing fluorescent reporters in BWM cells, we found that exopher formation (exophergenesis) is sex-specific and fertility-dependent. Moreover, exophergenesis is regulated by the developing embryo in utero, and exophers serve as transporters for muscle-generated yolk proteins, which can be used to nourish the next generation. Given the specific regulation of muscular exophergenesis, and the fact that muscle-generated exophers are much larger than neuronal ones and have different targeting, their identification and quantification required a modified approach from that designed for neuronal-derived exophers (Arnold et al., 2020). Here, we present a methodology for assessing and quantifying muscle-derived exophers that can be easily extended to determine their function and regulation in various biological contexts.
Graphical abstract

Background
Maintaining protein homeostasis (proteostasis) requires the degradation of damaged or unwanted proteins and plays a key role in the function of cells and organisms. The main proteolytic component of the cellular proteostasis network is the ubiquitin-proteasome system (UPS), by which protein substrates are labelled by attachment of a small ubiquitin protein and then targeted for degradation by the proteasome. In addition, the autophagy-lysosome pathway promotes proteostasis through the turnover of aggregated proteins and obsolete organelles (Mizushima and Komatsu, 2011; Gatica et al., 2018). Recently, a novel proteostasis mechanism was described in Caenorhabditis elegans , where protein aggregates, mitochondria, and lysosomes are removed from worm neurons to the hypodermis via large vesicles (of approximately 4 μm in diameter) called exophers (Melentijevic et al., 2017). Exophers are generated independently of the endosomal sorting complexes required for transport (ESCRT) machinery, and their surface lacks phosphatidylserine, distinguishing them from exosomes or apoptotic bodies. Conditions that interfere with proteostasis, and when protein turnover or autophagy is inhibited, increase exopher production. Exophers are sorted in the ced-1, ced-6, and ced-7 engulfment pathways (Melentijevic et al., 2017). Exophergenesis is evolutionarily conserved, as the malfunctioning mitochondria are excreted via exophers by mouse cardiomyocytes (Nicolás-Ávila et al., 2020), and exophers were identified in the human and mice neurons (Siddique et al., 2021). However, the biological function of exophers is not limited to eliminating cellular waste. We have demonstrated that the body wall muscle (BWM) of C. elegans can expel cellular content via robust exophers, which is later used to nourish the next generation. We identified these using the worms’ strains with fluorescently labeled mitochondria (GFP anchored in the mitochondrial membrane through a sequence of 50 aa of TOMM-20) and proteasome subunits, RPN-5 (19S subunit), PAS-7, or PAS-4 (20S subunits), as we initially studied exophers with regard to protein turnover. Muscle exophers, like their neuronal counterparts, are jettisoned from the cell body. Some remain connected to the extruding cell by a flexible tube that permits the transfer of cellular material to the attached vesicle (Figure 1A). Muscle exophers can also contain mitochondria, which in electron microscopy (EM) images showed increased surface area and disrupted cristae organization, as in cardiac exophers. Moreover, muscular exophergenesis is also not active during the larval stages, and its maximum level is reached around the second and third day of hermaphrodite adulthood (Turek et al., 2021). In addition, its activity is also reduced by the depletion of NADPH-cytochrome P450 reductase EMB-8 and actin-binding protein POD-1 (Melentijevic et al., 2017). However, in contrast to neurons or cardiomyocytes, we observed a low number of mitochondria-containing vesicles and a lack of significant changes in exophergenesis in response to proteotoxic stress. Our further analyses showed that BWM exophers represent a transgenerational metabolic/resource management system induced by the appearance of developing embryos in utero. In response, the newly formed exophers transport muscle-generated yolk proteins that can support the development of the offspring. Our data also suggests that BWM exophers are controlled by signals from developing embryos (Turek et al., 2021). Given the above, and that BWM exophers are generally more prominent in size than neuronal ones (up to 15 µm in diameter) and extruded into the body cavity, their identification, quantification, and analysis requires a modified methodology from that described for exophers of neuronal origin (Arnold et al., 2020). Here we provide details on the strains of C. elegans , time and conditions of growth, and a step-by-step procedure for imaging and quantifying BWM exophers. We believe this protocol will allow other laboratories to easily adapt their resources to decipher the autonomous and non-autonomous regulation of the BWM exophergenesis.
Materials and Reagents
Pipettes and pipette tips (Eppendorf)
1.5 mL and 50 mL polypropylene conical tube (VWR)
Fluorescent reporters produced from body wall muscle promoter myo-3 that label cytoplasmic proteins, and/or organelles (mitochondria or lysosomes) that will be excreted in exophers can be used to visualise and measure the exophergenesis. We commonly use ACH93 or ACH81 worm strains, where wrmScarlet is fused to proteasome subunit (RPN-5), and GFP is tagged to the TOMM-20 mitochondrial protein of the outer membrane, allowing for exopher visualization (wrmScarlet fluorescent signal) and inspection of mitochondria presence (GFP fluorescent signal) (Figure 1B–1C). We have prepared the following protocol for the C. elegans strains we use most often (Table 1), but this procedure will also be effective for other lines, as, those based on cytoplasmic GFP produced in BWM (Pmyo-3::GFP), which is also present in released exophers.
Table 1. Strains we use to study muscle-derived exophers
C. elegans strain Description Source ACH93 wacIs1[myo-3p::rpn-5 CAI = 0.97::GGGGS Linker-wrmScarlet::unc-54 3′UTR, unc-119(+)], wacIs14[myo-3 promoter::tomm-20_1–50aa::attB5::mGFP::unc-54-3′UTR, unc-119(+)] Turek et al. (2021) ACH81 wacIs1[myo-3 promoter::rpn-5 CAI=0.97::Optimal Linker-wrmScarlet::unc-54 3’UTR, unc-119(+)] Turek et al. (2021) 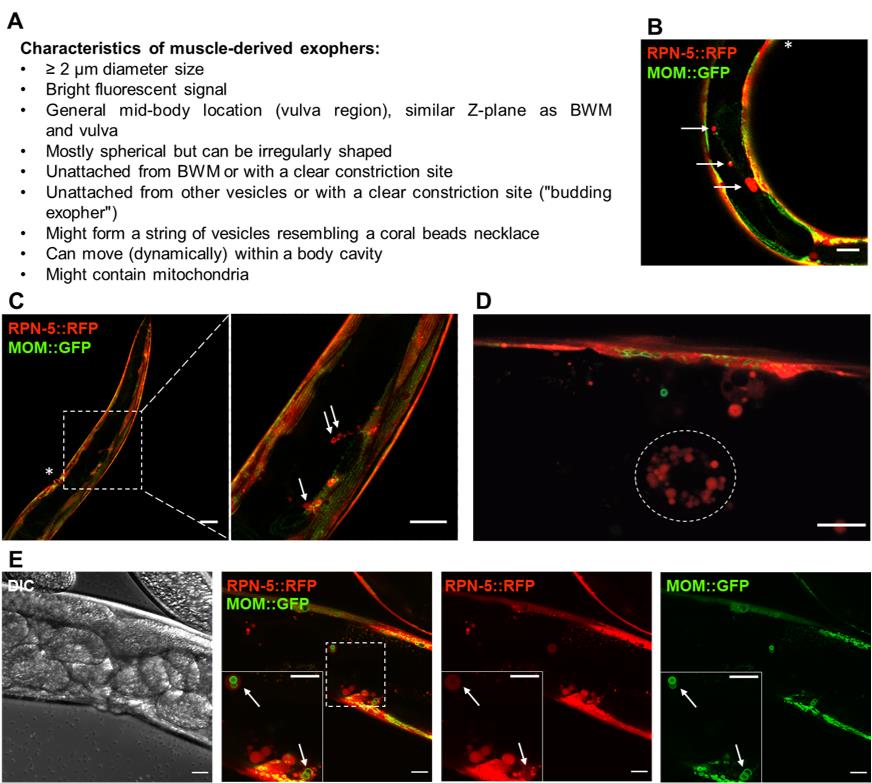
Figure 1. Fluorescently labelled (wrmScarlet) exophers extruded from the body wall muscles on day 2 of adulthood. A. Characteristics of muscle-derived exophers in C. elegans . B–C. Cross-sections of ACH93 worms (with a focus on the midbody) visualized in the RFP and GFP channels of the confocal microscope (Zeiss LSM800). The square region on panel C marked with a dashed line is magnified 3.2× on its right side. The arrows point to exophers, and the asterisks indicate the position of the vulva. Structures of an irregular, non-round shape or inside the BWM are not classified as exophers. Scale bars are 20 μm in B and 50 and 20 μm in C. D. The wrmScarlet signal may also originate from scattered and smaller vesicles (< 2 μm) present in the body cavity (circled with a dotted line). In our readings, we do not count these structures as exophers, as these are coelomocytes that degrade fluorescent proteins taken up from a pseudocoelom. Scale bar is 10 µm. E. Zoom into the vulva region in separate channels: DIC, a merge of RFP and GFP, separate RFP and GFP. The square marked with a dashed line marks the boundaries of the enlarged region in the bottom left corner. The arrows indicate exophers with mitochondria. MOM—mitochondrial outer membrane. Scale bar is 20 µm.LB broth (Sigma, catalog number: L3022-1KG)
Na2HPO4 (Sigma-Aldrich, catalog number: S7907)
KH2PO4 (Roth, catalog number: 3904.1)
NaCl (Chempur, catalog number: 117941206)
MgSO4 (Sigma-Aldrich, catalog number: M5921)
Peptone (BioShop, catalog number: PEP403.1)
Agar (BioShop, catalog number: AGR001.1)
KPO4 (Roth, catalog number: P749.1)
Streptomycin (Sigma-Aldrich, catalog number: S6501)
Nystatin (Sigma-Aldrich, catalog number: N1638)
Tetramisole hydrochloride (Sigma-Aldrich, catalog number: L9756)
Escherichia coli OP50 strain or any other bacteria that can serve as a food source (obtained from the Caenorhabditis Genetics Center, CGC) (see Recipes)
M9 buffer (see Recipes)
Nematode growth medium (NGM) (see Recipes)
Equipment
Fluorescence stereo or confocal microscope that allows for at least 200× magnification (ZEISS Axio Zoom.V16 Motorized Fluorescence Stereo Zoom Microscope)
Incubator for C. elegans (Q-Cell, PolLab)
Platinum home-made wire worm pick
Alcohol burner (DWK Life Sciences Wheaton)
Petri dishes for C. elegans cultivation (VWR)
Software
GraphPad Prism 9.3.1 (GraphPad Software)
Excel 2019 (Microsoft Office)
ZEN (Zeiss)
Procedure
文章信息
版权信息
© 2023 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Banasiak, K., Turek, M. and Pokrzywa, W. (2023). Preparation of Caenorhabditis elegans for Scoring of Muscle-derived Exophers. Bio-protocol 13(1): e4586. DOI: 10.21769/BioProtoc.4586.
分类
发育生物学 > 繁殖
细胞生物学 > 细胞成像 > 荧光
分子生物学 > 蛋白质 > 蛋白穿梭
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










