- EN - English
- CN - 中文
BONCAT-based Profiling of Nascent Small and Alternative Open Reading Frame-encoded Proteins
基于 BONCAT 的新生小和替代开放阅读框架编码蛋白质的分析
(*contributed equally to this work) 发布: 2023年01月05日第13卷第1期 DOI: 10.21769/BioProtoc.4585 浏览次数: 2128
评审: ASWAD KHADILKARThomas Farid MartínezMarie A Brunet

相关实验方案

高灵敏且可调控的 ATOM 荧光生物传感器:用于检测细胞中蛋白质靶点的亚细胞定位
Harsimranjit Sekhon [...] Stewart N. Loh
2025年03月20日 2270 阅读
Abstract
RIBO-seq and proteogenomics have revealed that mammalian genomes harbor thousands of unannotated small and alternative open reading frames (smORFs, <100 amino acids, and alt-ORFs, >100 amino acids, respectively). Several dozen mammalian smORF-encoded proteins (SEPs) and alt-ORF-encoded proteins (alt-proteins) have been shown to play important biological roles, while the overwhelming majority of smORFs and alt-ORFs remain uncharacterized, particularly at the molecular level. Functional proteomics has the potential to reveal key properties of unannotated SEPs and alt-proteins in high throughput, and an approach to identify SEPs and alt-proteins undergoing regulated synthesis should be of broad utility. Here, we introduce a chemoproteomic pipeline based on bio-orthogonal non-canonical amino acid tagging (BONCAT) (Dieterich et al., 2006) to profile nascent SEPs and alt-proteins in human cells. This approach is able to identify cellular stress-induced and cell-cycle regulated SEPs and alt-proteins in cells.
Graphical abstract
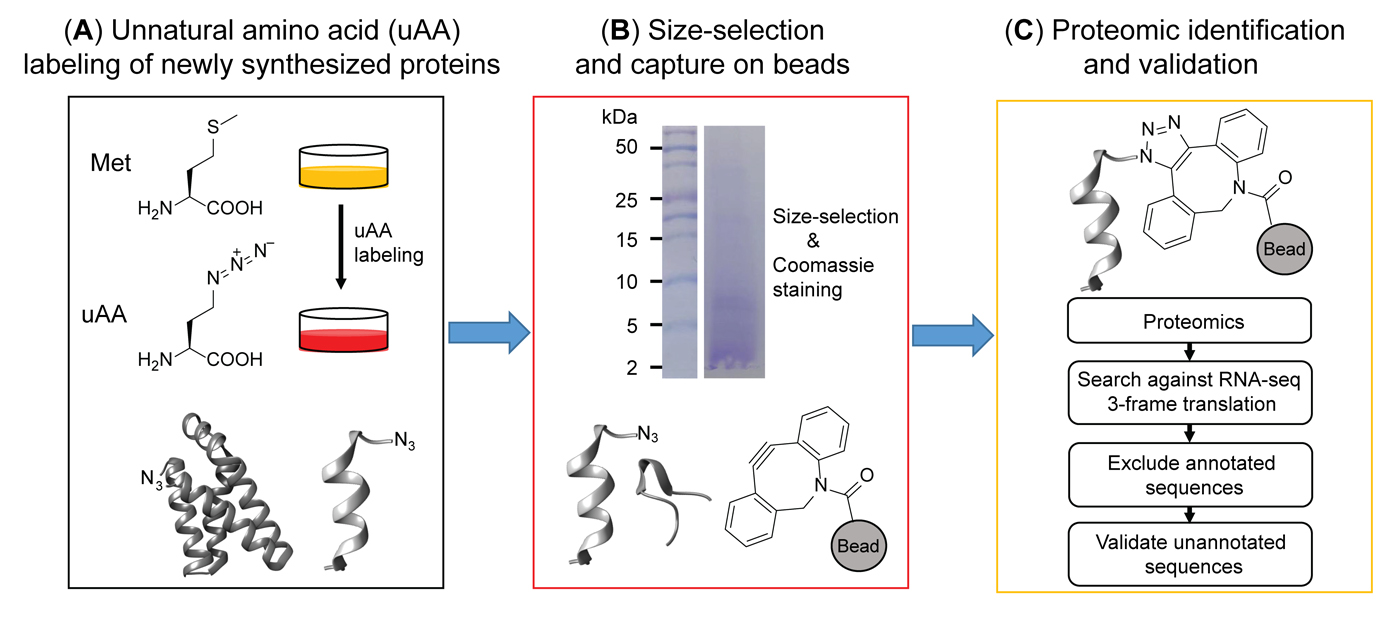
Schematic overview of BONCAT-based chemoproteomic profiling of nascent, unannotated small and alternative open reading frame-encoded proteins (SEPs and alt-proteins)
Background
Thousands of previously unannotated, expressed small open reading frames (smORFs) and alternative open reading frames (alt-ORFs) have recently been identified in mammalian genomes (Orr et al., 2020). These smORFs and alt-ORFs are found in 5' and 3' untranslated regions of mRNAs, in frame-shifted ORFs overlapping protein-coding sequences, and within long noncoding RNAs (including pseudogenes and antisense RNAs) (Brunet et al., 2018). Some smORF-encoded proteins (SEPs, also termed micropeptides or microproteins) and alt-ORF-encoded proteins (alt-proteins), which we will collectively refer to as “small proteins” in this protocol (Orr et al., 2020), have been shown to play important biological roles (Chen et al., 2020; Cao et al., 2021; Magny et al., 2021), suggesting that defining the functions of small proteins represents a major opportunity to gain insights into biology. However, due to their short lengths and limited homology to protein domains of known function, the majority of unannotated small proteins remain uncharacterized.
Similar to annotated proteins, the expression of some unannotated small proteins is cell-type specific (Cao et al., 2020), and is regulated by cellular stress (Jackson et al., 2018; Zhang et al., 2022). However, many prior reports on the discovery of unannotated small proteins do not provide information about differential expression or regulation. Methods to detect small proteins that are regulated by (patho)physiological processes and cellular stress should be of broad utility to enable hypothesis generation about their functions in high throughput.
Bio-orthogonal non-canonical amino acid tagging (BONCAT) incorporates the methionine analog azidohomoalanine (AHA) into all cellular proteins synthesized by the endogenous protein translation machinery during the labeling window (Dieterich et al., 2006). The first reported BONCAT workflows derivatize AHA-labeled proteins with biotin-alkyne, requiring a column-based step to remove excess biotin-alkyne, which de-enriches small proteins prior to streptavidin capture of the labeled proteome (Cao et al., 2022). Here, we describe a modified approach to profile nascent small proteins, which interfaces AHA labeling with in-solution size selection, followed by click chemistry capture directly on dibenzocyclooctyne magnetic beads, enabling sensitive detection of small proteins using mass spectrometry proteomics coupled with bioinformatic methods for unannotated protein identification. In a proof-of-principle study demonstrating this approach, we identified 22 actively translated, unannotated small proteins and N-terminal extensions of canonical proteins, in a cultured human cell line under control and stress conditions; we further confirmed that one of these unannotated small proteins is post-transcriptionally upregulated by DNA damage stress, and another one is cell-cycle-regulated (Cao et al., 2022), suggesting that this method may be broadly useful to reveal the regulated synthesis of unannotated small proteins in cultured cells.
Materials and Reagents
15-cm cell culture dish (Falcon, catalog number: 353025)
6-well cell culture plate (Greiner Bio-One, catalog number: 657160)
15 mL tube (Falcon, catalog number: 352096)
PolyWAX LPTM column (PolyLC, catalog number: 104WX0510)
Dulbecco’s modified Eagle’s medium (DMEM) (Corning, catalog number: 10-013-CV)
DMEM without methionine (DMEM-Met) (Corning, catalog number: 17-204-CI)
L-Azidohomoalanine (AHA) (Click Chemistry Tools, catalog number: 1066-100)
Fetal bovine serum (FBS) (Sigma, catalog number: F4135)
Penicillin/streptomycin (Pen/Strep) (Gibco, catalog number: 15140-122)
Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) (Click Chemistry Tools, catalog number: 1061-100)
Copper(II) sulfate (CuSO4) (Sigma, catalog number: C1297)
Biotin-alkyne (PEG4 carboxamide-Propargyl Biotin) (Click Chemistry Tools, catalog number: 1266-5)
Bond Elut C8 cartridge (Agilent, catalog number: 12105028)
DBCO (dibenzocycloctyne) magnetic beads (Click Chemistry Tools, catalog number: 1037-1)
2-Iodoacetamide (TCI, catalog number: I0741, CAS 144-48-9)
Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (Sigma, catalog number: C4706)
Sequencing grade modified trypsin (Promega, catalog number: V5111)
Ethyl acetate (Sigma, catalog number: 270989)
C18 spin columns (Thermo Scientific, catalog number: 89873)
Triethylamine (TEA) (Sigma, catalog number: 471283)
88%–91% formic acid (FA) (Sigma, catalog number: 399388)
Acetonitrile (ACN) (Sigma, catalog number: 271004)
Trifluoroacetic acid (TFA) (Sigma, catalog number: 302031)
Methanol (Sigma, catalog number: 34860)
Chloroform (Avantor, catalog number: JT-9180-01)
Triton X-100 (Amresco, catalog number: 0694)
DMSO (Dimethyl sulfoxide) (Alfa Aesar, catalog number: 36480)
Streptavidin-HRP (Invitrogen, catalog number: S911)
Ammonium bicarbonate (NH4HCO3) (Sigma, catalog number: A6141)
Urea (CH4N2O) (Sigma, catalog number: U5128)
Potassium chloride (KCl) (Avantor, catalog number: 3040-01)
Sodium carbonate (Na2CO3) (Sigma, catalog number: 230952)
Calcium chloride (CaCl2) (MP Biomedicals, catalog number: 153502)
Lysis buffer (see Recipes)
0.25 M TEAF pH 3.0 (see Recipes)
RIPA buffer (see Recipes)
4× SDS loading buffer (see Recipes)
Water-saturated ethyl acetate (see Recipes)
Equipment
Refrigerated centrifuge (Eppendorf, model: 5424 R)
Refrigerated centrifuge (Eppendorf, model: 5810 R)
SpeedVac vacuum concentrator (Thermo Scientific, Savant SPD10)
Vortex mixer (VWR, G-560 Vortexer 2)
Tube rotator (Thermo Scientific, catalog number: 05-450-127)
Heat block (Thermo Scientific)
37 °C incubator (Thermo Scientific)
Q Exactive Plus Hybrid Quadrupole Orbitrap (Thermo Scientific)
-80 °C freezer (Thermo Scientific)
-20 °C freezer (Thermo Scientific)
NEB magnetic holder (NEB, S1509S)
Agilent 1100 HPLC (Agilent)
Additional reagents and equipment for standard immunoblotting procedures
Software
Thermo Scientific Xcalibur (version 4.4)
Mascot (version 2.5.1)
Image Lab (version 5.2)
Agilent Chemstation for LC systems (version B.04.03)
Procedure
文章信息
版权信息
© 2023 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Cao, X., Chen, Y., Khitun, A. and Slavoff, S. A. (2023). BONCAT-based Profiling of Nascent Small and Alternative Open Reading Frame-encoded Proteins. Bio-protocol 13(1): e4585. DOI: 10.21769/BioProtoc.4585.
分类
分子生物学 > 蛋白质 > 检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











