- EN - English
- CN - 中文
Quantitative Analysis of Gene Expression in RNAscope-processed Brain Tissue
RNAscope 处理的脑组织中基因表达的定量分析
发布: 2023年01月05日第13卷第1期 DOI: 10.21769/BioProtoc.4580 浏览次数: 3080
评审: Arnau Busquets-GarciaFanny EhretAnonymous reviewer(s)
Abstract
Molecular characterization of different cell types in rodent brains is a widely used and important approach in neuroscience. Fluorescent detection of transcripts using RNAscope (ACDBio) has quickly became a standard in situ hybridization (ISH) approach. Its sensitivity and specificity allow for the simultaneous detection of between three and forty-eight low abundance mRNAs in single cells (i.e., multiplexing or hiplexing), and, in contrast to other ISH techniques, it is performed in a shorter amount of time. Manual quantification of transcripts is a laborious and time-consuming task even for small portions of a larger tissue section. Herein, we present a protocol for creating high-quality images for quantification of RNAscope-labeled neurons in the rat brain. This protocol uses custom-made scripts within the open-source software QuPath to create an automated workflow for the careful optimization and validation of cell detection parameters. Moreover, we describe a method to derive mRNA signal thresholds using negative controls. This protocol and automated workflow may help scientists to reliably and reproducibly prepare and analyze rodent brain tissue for cell type characterization using RNAscope.
Graphical abstract

Background
RNAscope is a powerful technology that allows for the identification of thousands of RNA targets visualized as puncta, each representing a single RNA transcript, in multiple types of tissues. Use of this assay has steadily increased since its introduction, allowing researchers across biological disciplines to perform rapid and precise quantification of RNA transcripts using a highly sensitive assay. In the neuroscience field, RNAscope is becoming an essential tool. Despite several publications in the field utilizing this technique, there is no standard methodology for processing brain tissue and quantifying RNA transcripts and transcript-positive cells using RNAscope. Many publications provide limited information regarding the calculation of quantification thresholds, preventing replication of prior analyses. Here, we outline a standardized protocol that can be used to establish criteria for quantifying cells that are positive for targets of interest using RNAscope, hopefully allowing for consensus across studies and improving reproducibility across the field. There are many ways in which RNAscope assays and analyses can be conducted; the purpose of this protocol is not to define a correct way, but rather to provide one potential methodology that can be used going forward and, perhaps more importantly, to start a conversation regarding unification and standardization of this approach across neuroscience labs.
Manual quantification of labeled cells is a tedious and time-consuming process. Many available tools used for image analysis (e.g., ImageJ) are often not suitable for analysis of large image files produced in RNAscope studies, and some open-source tools (e.g., dotdotdot or Cell Profiler) require advanced computational skills to run the workflow, thereby limiting their use. Here, we explain how to analyze RNAscope signal in rat neurons using the open-source bioimage quantification software QuPath (Bankhead et al., 2017). This software includes several features such as 1) annotation and visualization tools using a JavaFX interface, 2) built-in algorithms for common tasks, such as cell detection, and 3) interactive machine learning. Our method can be utilized for whole brain quantitative analysis or for analysis in selected brain regions as we performed in Avegno et al. (2021). Although we describe all the steps to generate RNAscope tissue from fresh frozen brains, the same method can be applied to fixed brain preparations with some modifications.
Here, we present the optimization of just one specific parameter important for automated cell detection, the fluorescence intensity threshold. However, we provide the framework for adding optimization steps for additional cell detection parameters, if desired. Collectively, the method presented here offers a user-friendly and cost-effective framework for automated quantification of transcript-positive cells in whole tissue sections, without the need for manual counting.
Materials and Reagents
Tissue Preparation
Rodent brains (we used n = 6 adult brains from female Wistar rats purchased from Charles River, catalog number: 003)
Isoflurane, USP, or any other anesthetic (Piramal Critical Care, catalog number: 6679401710; or any other brand)
250 mL glass beaker (Fisherbrand, catalog number: FB101250; or any brand)
Aluminum foil (Reynold Wraps, catalog number: 458742928317; or any brand)
Ultralow temperature digital thermometer with stainless-steel probe (Fisherbrand, catalog number: 15-077-32)
2-methylbutane (isopentane) ReagentPlus® ≥99% (Sigma-Aldrich, catalog number: M32631; or any brand)
Dry ice pellets
Cryostat (Thermo Scientific, USA, Cryostar NX50; or any brand capable of producing 10 µm slices)
Fisherbrand Superfrost Plus microscope slides (Fisher Scientific, catalog number: 12-550-15)
Tissue-Tek O.C.T. compound (Sakura, catalog number: 4583)
Tissue-Tek manual slide staining set (Sakura, catalog number: 4451)
Paraformaldehyde 32% aqueous solution (Electron Microscopy Sciences, catalog number: 15714S)
Distilled water
Fisherbrand Superslip coverslips (Fisher Scientific, catalog number: 12-545-89P)
Fluoro-Gel II with DAPI (Electron Microscopy Sciences, catalog number: 17985-50)
RNAscope Preparation
RNAscope® Fluorescent Multiplex reagent kit v1 (Advanced Cell Diagnostics, catalog number: 320850) for fresh frozen applications. Note that for fixed brain preparations, additional products are needed: RNAscope Target Retrieval and RNAscope Protease III (available in the RNAscope Universal Pretreatment kit, Advanced Cell Diagnostics, catalog number: 322380)
RNAscope® RTU Protease IV reagent (Advanced Cell Diagnostics, catalog number: 322340)
RNAscope target probes (catalog number varies depending on the need; we used Rn-Hcrtr1-C1, Rn-Th-C2, and Rn-Fos-C3). Note that in order to independently detect mRNA transcripts in a multiplex assay, each target probe must be in a different channel (C1, C2, or C3) and one of the target probes must be in the C1 channel. Channel C1 target probes are ready-to-use (RTU), while channel C2 and C3 probes are shipped as a 50× concentrated stock. The 50× probes for C2 or C3 must be mixed with a C1 RTU probe. If no specific C1 probe is used, then a blank probe diluent (assay dependent) is used to dilute the probes. Assignation of channels can be changed upon researcher request.
RTU probe diluent (optional, see above; Advanced Cell Diagnostics, USA, catalog number: 300041)
RNAscope 3-plex negative control probes (Advanced Cell Diagnostics, catalog number: 320871)
Immedge® hydrophobic barrier pen (Vector Labs, catalog number: 310018)
RNase AWAY (Thermo Scientific, catalog number: 14-375-35) or any other decontaminant (e.g., bleach)
Notes:
The items listed below are intended for fresh frozen tissue collection only. If researchers plan to use formalin-fixed tissue, there will be additional steps to be followed (refer to manufacturer manuals for more details).
RNAscope kits for fresh frozen and fixed tissues are slightly different, as the fixed tissue follows a different preparation and pretreatment.
Target probes listed here are merely indicative. Any type of marker can be utilized for this purpose. Customized probes (from available genome databases) can be made upon request at additional costs.
Equipment
Brain extraction tools:
Straight scissors (Roboz, catalog number: RS-6806), rongeurs (Roboz, catalog number: RS-8300), rounded spatula (Millipore Sigma, catalog number: HS15906), small straight or curved forceps (Roboz, catalog number: RS-5136), and long forceps (Fisherbrand, catalog number: 13-820-077; see Figure 1)
Slide scanner (Carl Zeiss, AxioScan Z.1, Germany; or any other microscope capable of scanning and digitizing fluorescent slides)
Computer: 64-bit Windows or Linux with minimum 16 GB RAM and a fast multicore processor (e.g., Intel Core i7 or i9)
HybEZ II system hybridization oven [Advanced Cell Diagnostics, USA; see Figure 2. The system comprises: HybEZ oven (PN 321710/321720), a humidity control tray (PN 310012), and HybEZ humidifying paper (2 sheets, PN 310025), EZ-Batch wash tray (PN 321717), and EZ-Batch slide holder (PN 321716)]
Note: This protocol will prospectively work with a less powerful computer or with lower RAM, but analysis will be slow and may encounter memory errors.
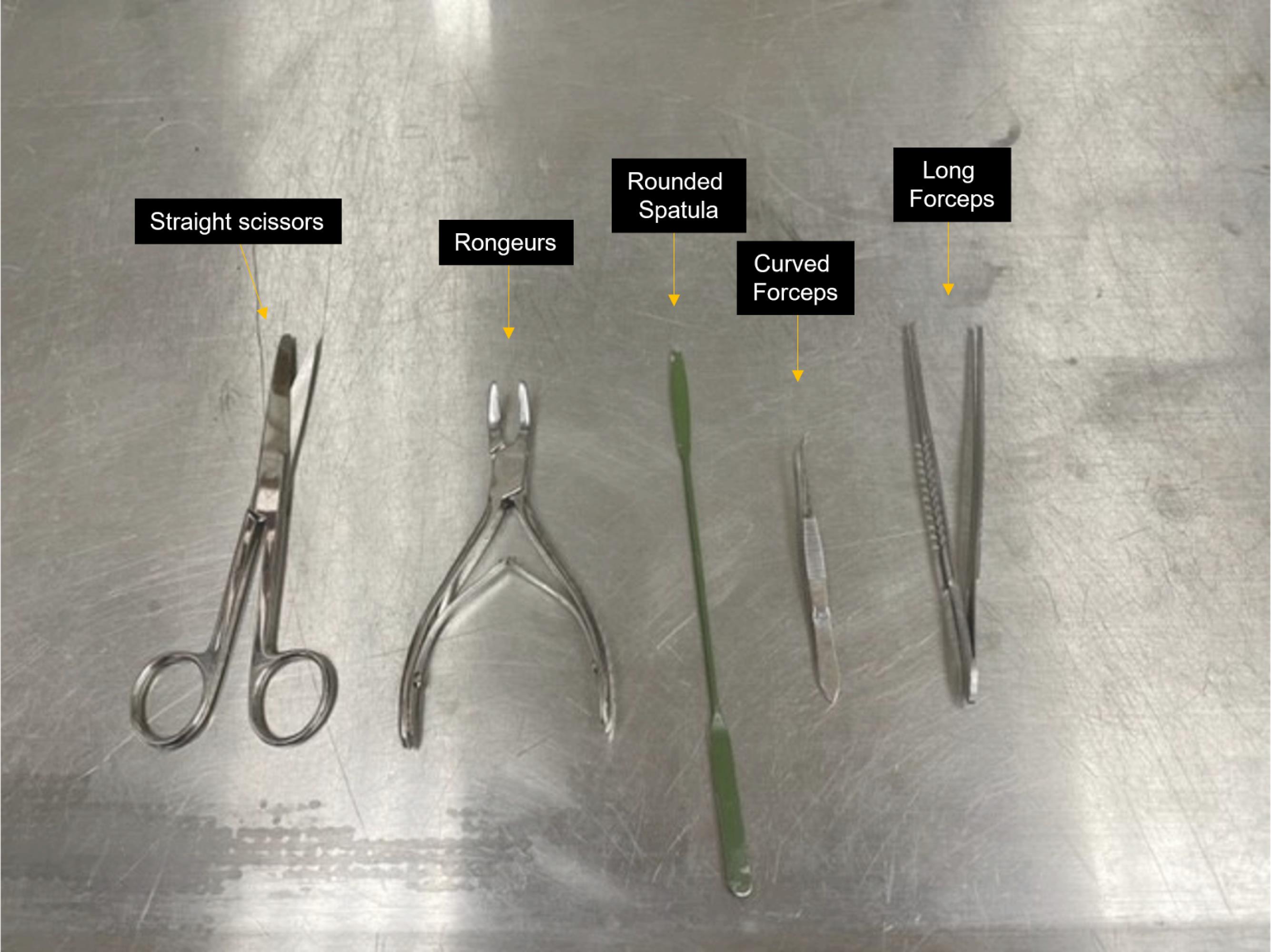
Figure 1. Brain extraction tools. Straight scissors, rongeurs, rounded spatula, and curved and long forceps indicated.
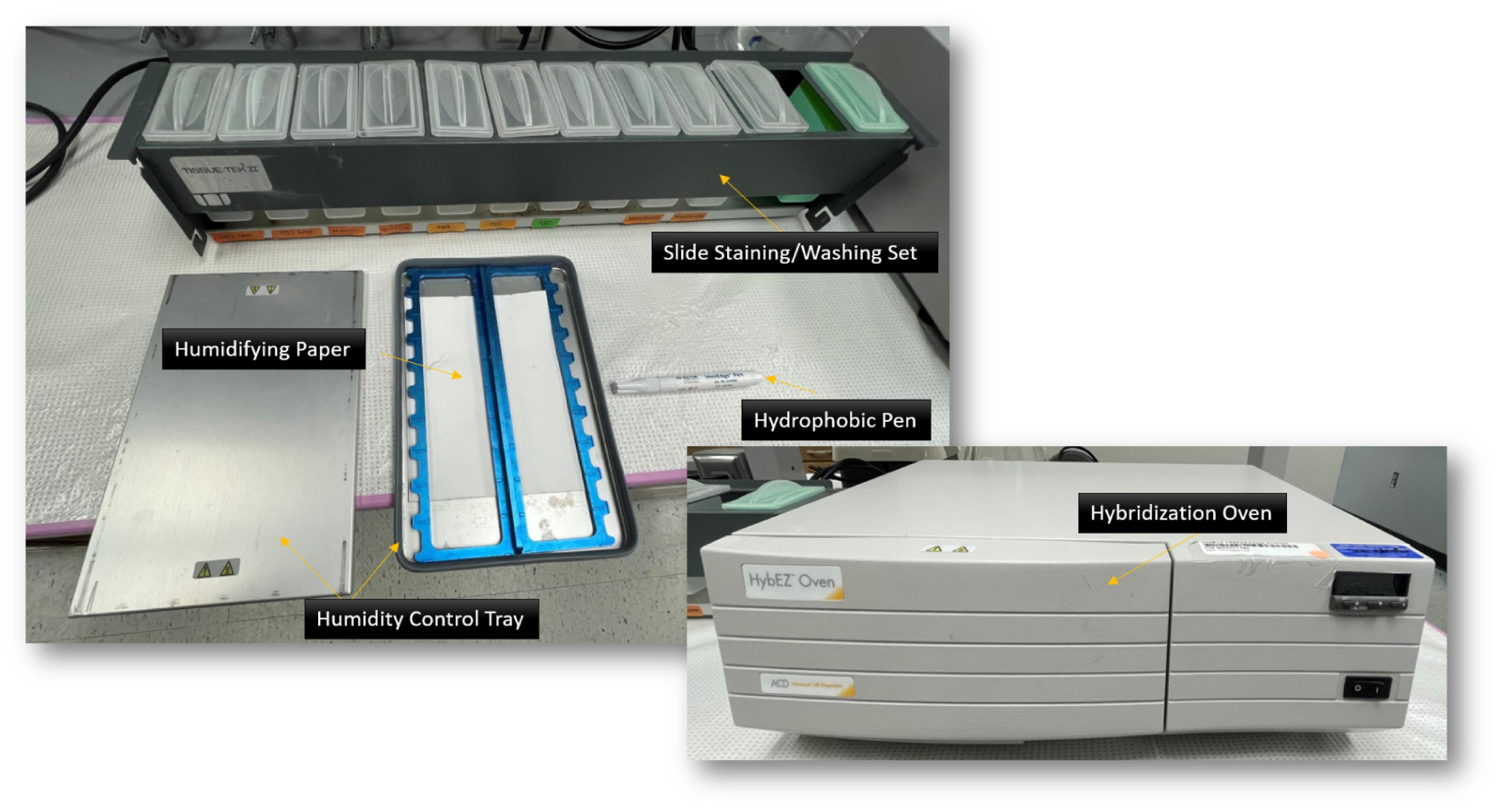
Figure 2. HybEZ II system. Hybridization oven, humidity control tray, humidifying paper, slide staining/washing set (EZ-Batch wash tray and EZ-Batch slide holder), and hydrophobic pen indicated.
Software
Zeiss ZEN blue v2.6 or higher for images acquisition and export (https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html#downloads)
QuPath 0.3.2 open-source software (https://qupath.github.io/)
Microsoft Excel or similar
GraphPad Prism 9.4.1 (https://www.graphpad.com/) or similar
Procedure
文章信息
版权信息
© 2023 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Secci, M. E., Reed, T., Quinlan, V., Gilpin, N. W. and Avegno, E. M. (2023). Quantitative Analysis of Gene Expression in RNAscope-processed Brain Tissue. Bio-protocol 13(1): e4580. DOI: 10.21769/BioProtoc.4580.
分类
神经科学 > 神经解剖学和神经环路 > 荧光成像
神经科学 > 基础技术
细胞生物学 > 基于细胞的分析方法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link