- EN - English
- CN - 中文
A Fluorescence-based Approach Utilizing Self-labeling Enzyme Tags to Determine Protein Orientation in Large Unilamellar Vesicles
利用自标记酶基于荧光方法确定大单层囊泡中的蛋白质定位
(*contributed equally to this work) 发布: 2022年11月05日第12卷第21期 DOI: 10.21769/BioProtoc.4542 浏览次数: 2325
评审: David PaulKumiko OkazakiAnonymous reviewer(s)
Abstract
Reconstitution of membrane proteins into large unilamellar vesicles is an essential approach for their functional analysis under chemically defined conditions. The orientation of the protein in the liposomal membrane after reconstitution depends on many parameters, and its assessment is important prior to functional measurements. Common approaches for determining the orientation of a membrane-inserted protein are based on limited proteolytic digest, impermeable labeling reagents for specific amino acids, or membrane-impermeable quenchers for fluorescent proteins. Here, we describe a simple site-specific fluorescent assay based on self-labeling enzyme tags to determine the orientation of membrane proteins after reconstitution, exemplified on a reconstituted SNAP-tag plant H+-ATPase. This versatile method should benefit the optimization of reconstitution conditions and the analysis of many types of membrane proteins.
Graphical abstract:
Background
Reconstitution of purified proteins into model membranes is an essential approach for investigating membrane protein function under defined conditions outside the complex cellular environment (Rigaud and Lévy, 2003; Murray et al., 2014; Amati et al., 2020). Typically, proteins are reconstituted with phospholipids into large unilamellar vesicles with 100–200 nm diameters. The orientation of the protein in the liposomal membrane after reconstitution depends on many parameters, including the type of protein, the lipid composition, and the reconstitution procedure (Tunuguntla et al., 2013; Amati et al., 2020). Knowledge of the orientation distribution of the protein is of crucial significance for interpreting its function in the proteoliposomal in vitro system and for the design of proteoliposome experiments. A common approach for determining the orientation of a membrane-inserted protein is the protease-mediated cleavage (e.g., Schuette et al., 2004; Serek et al., 2004; Islam et al., 2013; Eisinger et al., 2018). This assay is based on the principle that an externally added protease can cleave only the accessible portions of the membrane-inserted protein, which requires several critical steps such as limited proteolytic digest, stopping the proteolytic reaction, and fragment analysis (e.g., by SDS-PAGE).
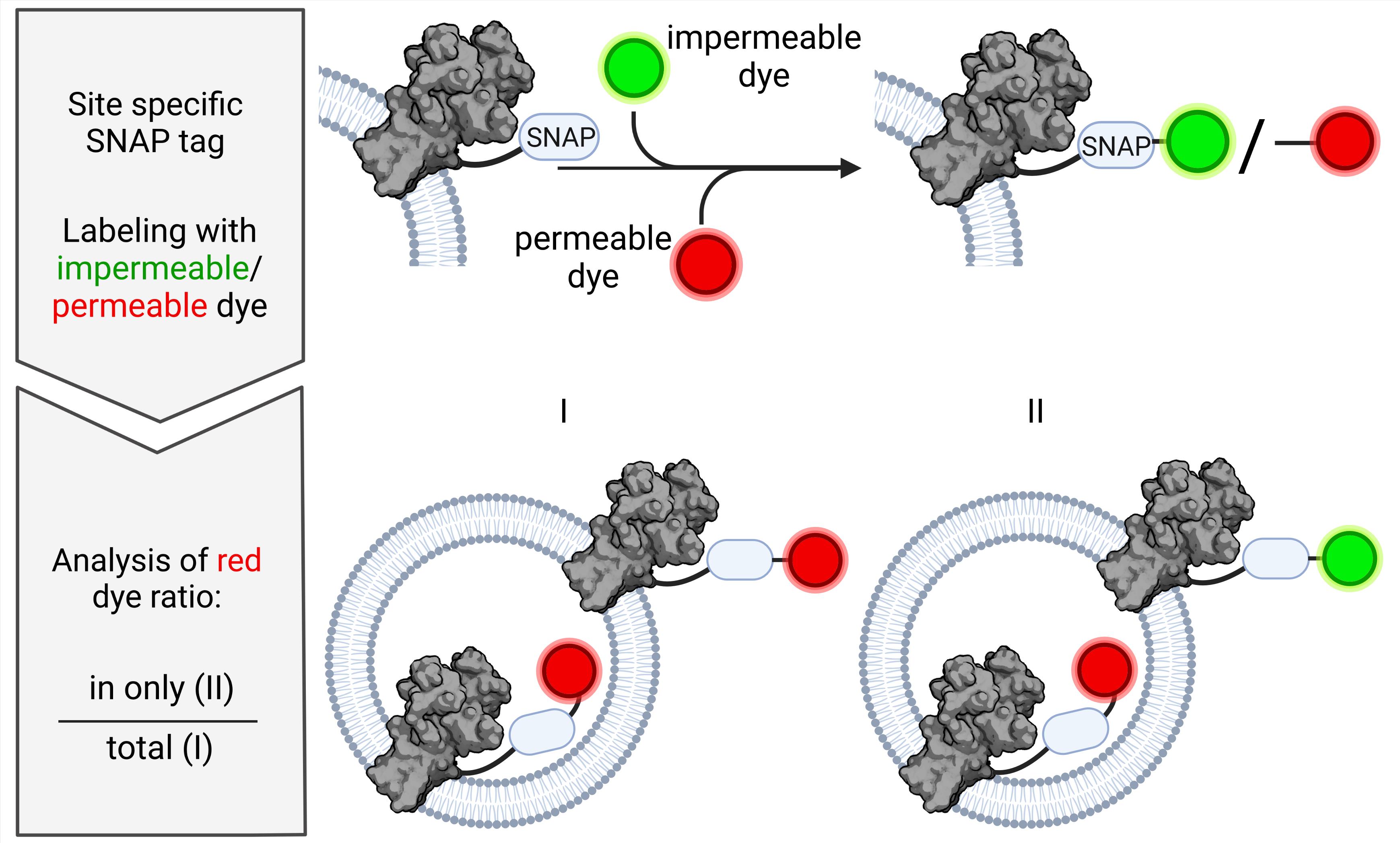
Alternative approaches are based on impermeable labeling reagents for specific amino acids or membrane-impermeable quenchers for fluorescent proteins (e.g., Zhang et al., 2003; Marek et al., 2011; Deutschmann et al., 2022). The protocol described here takes advantage of the SNAP-tag technology (Keppler et al., 2004; Cole, 2013), whereby a protein can be labeled site-specific after expression or purification with a diversity of dyes, which are either membrane-permeable or impermeable (Figure 1). This approach can be extended to other self-labeling enzyme tags including SNAP-tag, HALO-tag, or CLIP-tag and, thus, can be easily customized for the protein of interest. In contrast to other labels such as cysteine labeling, side-specific, stoichiometric 1:1 labeling without changing the amino acid sequence is ensured. Compared to protease-based approaches, labeling with dyes of different permeability allows analysis under nondestructive conditions for the vesicle and the protein. The presented strategy will be useful for the optimization of reconstitution conditions and the analysis of many types of membrane proteins. Thus, this new assay expands the toolbox of fluorescence-based methods for the estimation of membrane protein orientation in liposomes.

Figure 1. SNAP-labeling process. The membrane protein of interest is expressed with SNAP-tag enabling site-specific labeling. Based on the human DNA repair enzyme O6-alkylguanine-DNA-alkyltransferase, the SNAP enzyme undergoes self-labeling via a covalent bond with O6-benzylguanine derivatives. Depending on the characteristics of the derivative, the labeling substrate is either membrane-permeable like SNAP-647-SIR or membrane-impermeable like SNAP-Alexa488 [chemical structures based on Lukinavičius et al. (2013) and Wilhelm et al. (2021), and drawn with https://chem-space.com].
Things to consider before starting
Choice of labeling substrate
Here, we used a SNAP-tagged fused membrane protein and benzylguanine-derived substrates. Labeling of the SNAP-tag is irreversible and quantitative, and thus well suited for the detection and quantitation of labeled proteins via in-gel fluorescence scanning of SDS-PAGE gels. If alternative labeling systems based on, for example, CLIP-tag or ACP-tag, are used, the according substrate has to be chosen as basis for the labeling dye.
Choice of self-labeling fluorophore
The assay relies on the use of a membrane-impermeable dye in combination with a membrane-permeable dye. In addition, both dyes should be selected for low spectral overlap and based on the imaging system available.
Choice of lipid environment
Here, we describe labeling conditions optimized for proteins reconstituted in 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) / 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) (9:1 mol/mol) vesicles. In other lipid environments, labeling duration and label concentrations might need adjustment.
Additives in buffer
A reducing agent (e.g., DTT) is added to the labeling buffer to increase labeling efficiency, due to a reactive cysteine in the SNAP-tag. However, if its presence is affecting the protein, the experiment could be tested without DTT, with optimized labeling time and label concentrations. However, under oxidizing conditions, labeling might not be achieved efficiently. The presence of chelating reagents such as EDTA should be avoided as the SNAP-tag protein contains a structural Zn2+ ion.
Biological materials
The exemplary detergent-solubilized SNAP-tagged membrane protein is a C-terminally truncated version (∆73) of the Arabidopsis thaliana auto-inhibited H+-ATPase isoform 2 (AHA2-SNAP) with additional StrepII and hexahistidine N-terminal tags. AHA2-SNAP was heterologously expressed in the Saccharomyces cerevisiae strain RS-72 (MATa, ade1-100 his4-519 leu2-3,112; endogenous proton pump PMA1 gene is under control of GAL1 promoter) (Cid et al., 1987) and purified via the His-tag resulting in 5–10 mg/mL protein in storage buffer (see Recipe 3), stored at -80 °C (Lanfermeijer et al., 1998).
Note: The protocol can also be applied to purified membrane proteins prepared differently in combination with liposomal reconstitution procedures not based on preformed liposomes, as described here. However, these membrane proteins may have different preferences for lipids, with respect to the phospholipid headgroup and the lipid fatty acid composition.
Materials and Reagents
All catalog numbers provided below shall serve as guide; alternative sources can be used as well.
Materials
2 mL disposable syringe (Henry Schein, catalog number: 9003017)
Pipette tips 10 µL, 200 µL, and 1,000 µL (Sarstedt, catalog numbers: 70.760.002, 70.3030.020, and 70.3050.020)
Reaction tubes 1.5 mL, 2 mL, 15 mL, and 50 mL (Sarstedt, catalog numbers: 72.690.001, 72.691, 62.554.502, and 62.547.254)
Glass beads, 3 mm (Supelco, catalog number: 1040150500)
Round bottom glass tube (Roth, catalog number: NY90.1)
Screw cap, ND8 (Roth, catalog number: NL96.1)
Screw neck ND8 vial (Roth, catalog number: KE27.1)
Sterican disposable 26 gauge needle, 0.45 × 12 mm (Braun, catalog number: 4665457)
Chemicals
Chloroform, ethanol-stabilized and certified for absence of phosgene and HCl (Roth, catalog number: 7331.2)
Methanol (VWR, catalog number: 20847.307)
4-Morpholinepropanesulfonic acid, 3-(N-Morpholino)propanesulfonic acid (MOPS) (Sigma, catalog number: M3183)
Dimethylsulfoxide (DMSO) (Sigma, catalog number: 34943)
KOH (Fisher Chemical, catalog number: P/5640/60)
K2SO4 (VWR, catalog number: 26997.293)
Sodium acetate (Sigma, catalog number: S2889)
2-(N-Morpholino)-ethane sulphonic acid (MES) (Roth, catalog number: 4256.2)
Glycerol (VWR, catalog number: 24388.295)
KCl (Honeywell, catalog number: 31248)
Ethylenediaminetetraacetic acid (EDTA) (Sigma, catalog number: E6758)
threo-1,4-dimercapto-2,3-butanediol (DTT) (Sigma, catalog number: 43819)
Liquid nitrogen
N-dodecyl β-maltoside (DDM) (Glycon, catalog number: D97002)
N-octyl β-D-glucopyranoside (OG) (Glycon, catalog number: D97001)
Sephadex G50, fine (Cytiva, catalog number: 17004201)
BioBeads (Bio-Rad, catalog number: 152-3920).
Bromophenol blue (Roth, catalog number: A512.3)
Tris(hydroxymethyl)aminomethan (Tris) (Sigma, catalog number: T1503)
Sucrose (Fisher Chemical, catalog number: S/8600/60)
N,N,N′,N′-tetramethyl ethylenediamine (TEMED) (Merck, catalog number: 8.08742.0250)
Ammonium peroxydisulfate (APS) (Roth, catalog number: 9592.2)
Sodium dodecyl sulfate (SDS) (Sigma, catalog number: L3771)
Aluminum sulfate hydrate (Roth, catalog number: 3731.1)
Ethanol absolute ≥99.8% (VWR, catalog number: 20821.321)
Acrylamide (Roth, catalog number: 3029.1)
Orthophosphoric acid (VWR, catalog number: 20624.295)
Coomassie brilliant blue G-250 (Serva, catalog number: 35050)
PageRulerTM Plus Prestained Protein Ladder, 10–250 kDa (Thermo Scientific, catalog number: 26619)
Reconstitution buffer (see Recipes)
Protein storage buffer (see Recipes)
1 M OG stock (see Recipes)
0.5 M DTT stock (see Recipes)
Sephadex G50 fine slurry (see Recipes)
Pre-washed BioBeads (see Recipes)
Laemmli buffer (see Recipes)
20% SDS stock (see Recipes)
SDS-PAGE running buffer (see Recipes)
SDS gel (see Recipes)
10% APS stock (see Recipes)
1 M Tris (see Recipes)
Colloidal Coomassie staining solution (see Recipes)
Colloidal Coomassie destaining solution (see Recipes)
Lipids
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Avanti Polar Lipids, catalog number: 850457)
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) (POPG) (Avanti Polar Lipids, catalog number: 840457)
SNAP dyes in DMSO
SNAP-Surface Alexa488 (New England BioLabs Inc., catalog number: S9129S)
SNAP-Cell 647-SiR (New England BioLabs Inc., catalog number: S9102S)
Note: The assay has also been successfully performed using SNAP-Surface 647 (New England BioLabs Inc., catalog number: S9136S) and SNAP-Cell Oregon Green (New England BioLabs Inc., catalog number: S9104S)
Equipment
Analytical balance (Sartorius Entris-i II, 220 g/0.1 mg, Buch Holm, catalog number: 4669128)
Avanti mini extruder set (Avanti Polar Lipids, catalog number: 610000)
Filter supporter for extruder (Polyester Drain Disc, 10 mm; Cytiva, catalog number: 230300)
Membrane 200 nm for extruder (Cytiva, catalog number: 10417004)
1,000 μL gas-tight syringe (Avanti Polar Lipids, catalog number: 610017)
Centrifuge with rotor for 15 and 50 mL polypropylene tubes (Eppendorf 5810 R; Wesseling, Germany)
Chemidoc MP imaging system (Bio-Rad) with illumination at 460–490 nm and 520–545 nm with 530/28 nm and 695/55 nm emission filters, respectively.
Eppendorf Research® plus pipettes P2.5, P20, P200, and P1000 (Eppendorf, catalog numbers: 3123000012, 3123000039, 3123000055, and 3123000063)
Glass desiccator Boro 3.3 with socket in lid, 20 cm, including stopcock (BRAND GmbH, catalog number: 65238)
Hamilton 700 Series syringes 25 µL, 100 µL, and 1,000 µL (Hamilton Company, Nevada, USA)
Head-over-head turning device (neoLab, catalog number: 7-0045)
Mini-PROTEAN® Tetra cell system (Bio-Rad, catalog number: 1658000)
Vacuum Pump V-100 with interface I-100 and rotary evaporator Rotavapor® R-100, SJ29/32, V, 220–240V (Buchi, catalog numbers: 11593636, 11593655D and 11100V111, 11061895)
Vortex mixer (Scientific Industries Inc., model: Vortex Genie 2, catalog number: SI-0236)
Water bath (Julabo, CORIO C-BT5, catalog number: 9011305)
Software
ImageLab software version 5.2.1 (Bio-Rad)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Paweletz, L. C., Veit, S. and Pomorski, T. G. (2022). A Fluorescence-based Approach Utilizing Self-labeling Enzyme Tags to Determine Protein Orientation in Large Unilamellar Vesicles. Bio-protocol 12(21): e4542. DOI: 10.21769/BioProtoc.4542.
分类
生物工程 > 合成生物学
生物化学 > 蛋白质 > 荧光
生物物理学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link