- EN - English
- CN - 中文
CRISPR/Cas9-mediated LRP10 Knockout in HuTu-80 and HEK 293T Cell Lines
HuTu-80 和 HEK 293T 细胞系中 CRISPR/Cas9 介导的 LRP10 敲除
发布: 2022年10月05日第12卷第19期 DOI: 10.21769/BioProtoc.4521 浏览次数: 3646
评审: Xi FengAnonymous reviewer(s)

相关实验方案
利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1175 阅读
Abstract
Loss-of-function (LoF) variants in the low-density lipoprotein receptor–related protein 10 gene (LRP10) have been recently implicated in the development of neurodegenerative diseases, including Parkinson's disease (PD), PD dementia (PDD), and dementia with Lewy bodies (DLB). However, despite the genetic evidence, little is known about the LRP10 protein function in health and disease. Here, we describe a detailed protocol to efficiently generate a LRP10 LoF model in two independent LRP10-expressing cell lines, HuTu-80 and HEK 293T, using the CRISPR/Cas9 genome-editing tool. Our method efficiently generates bi-allelic LRP10 knockout (KO), which can be further utilized to elucidate the physiological LRP10 protein function and to model some aspects of neurodegenerative disorders.
Graphical abstract:
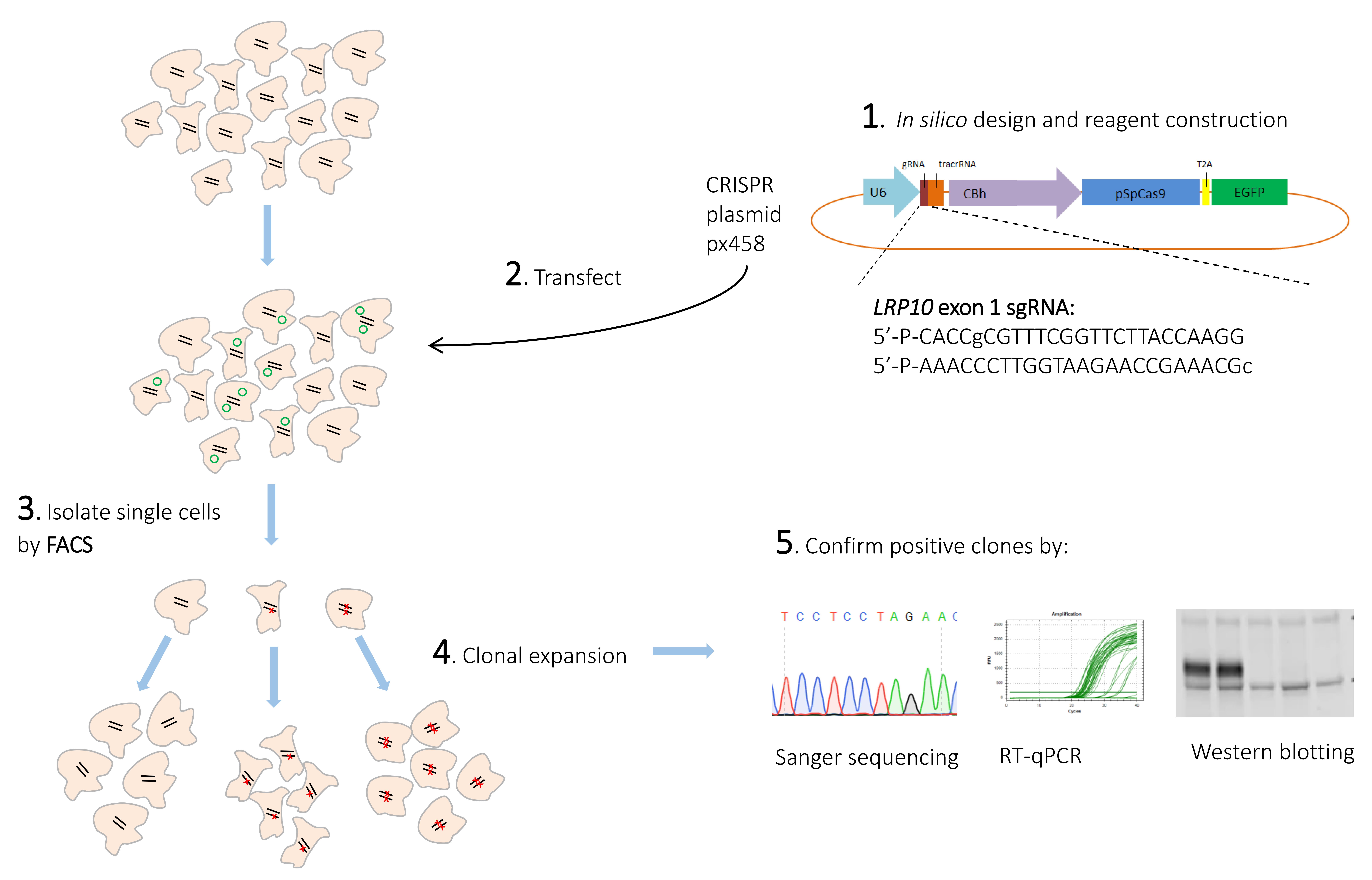
CRISPR/Cas9 workflow for the generation of the LRP10 KO. (1) Designed single guide RNA (sgRNA) is cloned into CRISPR/Cas9 px458 plasmid. (2) Cells are transfected with the CRISPR/Cas9 plasmid containing sgRNA. (3) Two days post transfection, cells are dissociated and sorted as single cells by fluorescence-activated cell sorting (FACS). (4) After several weeks, expanded clonal lines are (5) verified with Sanger sequencing for the presence of INDELs (insertions or deletions), RT-qPCR for the amounts of LRP10 mRNA transcript, and Western blotting for the analysis of the LRP10 protein levels.
Background
Rare, pathogenic variants in the low–density lipoprotein–related protein 10 gene (LRP10) have been identified in patients with familial Parkinson’s disease (PD), PD dementia (PDD), and dementia with Lewy bodies (DLB) (Quadri et al., 2018). Moreover, postmortem analysis of patients carrying distinct LRP10 variants showed a severe burden of alpha-synuclein–associated pathology in the form of Lewy bodies (LB) and Lewy neurites (LN) in the brain stem, limbic, and cortical regions (Quadri et al., 2018). Importantly, functional studies revealed that initially identified LRP10 pathogenic variants affected either LRP10 transcript expression and stability, protein stability, or protein localization, pointing to loss-of-function (LoF) as a shared pathogenic mechanism (Quadri et al., 2018). In addition, earlier studies using LRP10 overexpression models reported that LRP10 participates in intracellular trafficking pathways (Boucher et al., 2008; Brodeur et al., 2009, 2012; Doray et al., 2008). Despite these data, little is known about the endogenous LRP10 function in health and disease, partly due to the lack of in vitro LRP10 knockout (KO) models.
The CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 system is a powerful and precise method for editing the genome in various cell types (Jinek et al., 2012; Cong et al., 2013; Adli, 2018). Gene KO models are often critical for identifying the function of the protein that a particular gene encodes. In the CRISPR/Cas9 system, the endonuclease Cas9 is guided to a specific site in the genome by a single guide RNA (sgRNA) to generate a double-strand break (Adli, 2018). The break is fixed in a process called non-homologous end-joining (NHEJ), which results in INDELs (insertion or deletion) (Sander and Joung, 2014). The introduction of INDELs leads to changes in the reading frame, which disrupts mRNA and protein expression (Sander and Joung, 2014).
Here, we show a step-by-step protocol to efficiently generate a LRP10 KO in the epithelial human cell lines HuTu-80 and HEK 293T using the CRISPR/Cas9 genome-editing tool. Our approach efficiently targeted the first exon of the LRP10 gene using a sgRNA cloned into a plasmid containing Cas9 from Streptococcus pyogenes. We obtained homozygous and compound heterozygous independent LRP10 KO clones carrying INDEL mutations leading to a frameshift and a premature stop codon. These models were used to test the specificity of commercially available and in-house developed antibodies against the endogenously expressed LRP10 protein (Grochowska et al., 2021). Interestingly, HuTu-80 cells highly express both LRP10 and alpha-synuclein, making the LRP10 KO in HuTu-80 cells a suitable model for studying the potential link between LRP10 LoF and alpha-synuclein accumulation in PD, PDD, and DLB. Lastly, given the high targeting efficiency of the LRP10 locus in both cell lines, this protocol holds promise for the generation of LRP10 KO in more relevant in vitro models of neurodegeneration, such as human stem cell–derived neural progenitors that can be differentiated into neurons and glia.
Materials and Reagents
6-well cell culture plates (TC-plate, standard F, Sarstedt, catalog number: 83.3920.005)
48-well cell culture plates (TC-treated, F-bottom, CorningTM CostarTM, catalog number: 3548)
96-well cell culture plates (F-bottom with lid, Greiner Bio-One, catalog number: 655180)
FalconTM round-bottom polystyrene test tubes with cell strainer snap cap, 5 mL (FalconTM, catalog number: 352235)
10 cm Petri dishes (sterile, VWR)
Parafilm® M (Sigma-Aldrich, catalog number: P7793)
DMEM (Sigma-Aldrich, catalog number: D6429), store at 4 °C
DMEM/F-12 (GibcoTM, catalog number: 11320033), store at 4 °C
Fetal bovine serum (FBS, GibcoTM, catalog number: A5256701), aliquot and store at -20 °C
Trypsin–EDTA (0.05%), phenol red (GibcoTM, catalog number: 25300054), aliquot and store at -20 °C
Penicillin–streptomycin (10,000 U/mL, GibcoTM, catalog number: 15140122), aliquot and store at -20°C
DPBS, no calcium and no magnesium (DPBS-/-, GibcoTM, catalog number: 14190144), store at room temperature
pSpCas9-(BB)-2A-GFP (Addgene plasmid #48138; http://n2t.net/addgene:48138; RRID: Addgene_48138)
BbsI (10,000 units/mL; NEB, catalog number: R0539S), aliquot and store at -80 °C
NEBufferTM r2.1 (NEB, catalog number: B7030S), store at 4 °C
T4 DNA ligase (400,000 units/mL, NEB, catalog number: M0202S), store at -20 °C
T4 DNA ligase reaction buffer (NEB, catalog number: B0202S), store at -20 °C
TE buffer solution 1× (TRIS–EDTA buffer) pH 8,0 (VWR, J75793.AE), store at room temperature
Ampicillin (Sigma-Aldrich, 69-52-3)
BD BACTOTM agar (BD, catalog number: 214010)
Tryptone (Millipore, catalog number: T7293)
Yeast extract (Sigma-Aldrich, catalog number: Y1625)
NaCl (Sigma-Aldrich, catalog number: S7653)
Trizma® base (Sigma-Aldrich, catalog number: TRIS-RO)
cOmpleteTM (Roche, catalog number: 11836145001)
Pefabloc® SC (Roche, catalog number: 11585916001)
IGEPAL® CA-630 (Sigma-Aldrich, catalog number: 18896)
SDS (Sigma-Aldrich, catalog number: 71729)
Bromophenol blue (Thermo Fisher Scientific, catalog number: A18469.18)
Glycerol (Sigma-Aldrich, catalog number: G5516)
DTT (Dithiothreitol, Thermo Fisher Scientific, catalog number: R0862)
TWEEN® 20 (Sigma-Aldrich, catalog number: P1379)
Agarose powder (Sigma, catalog number: A9539)
GeneJuice® transfection reagent (Merck Millipore, catalog number: 70967), store at 4 °C
Hoechst 33342, trihydrochloride, trihydrate, 10 mg/mL solution in water (InvitrogenTM, catalog number: H3570)
One ShotTM TOP10 chemically competent E. coli (InvitrogenTM, catalog number: C404010), store at -80 °C
HuTu-80 cells (CLS, catalog number: 330218), expand and cryopreserve cell stocks in liquid nitrogen
HEK 293T (ATCC® CRL-3216TM), expand and cryopreserve cell stocks in liquid nitrogen
DMSO (Sigma-Aldrich, catalog number: W387509), aliquot and store at -20 °C
Sanger sequencing kit (BigDye Terminator v3.1 Cycle Sequencing Kit and ExoSAP-IT, A38073), store the components according to the manufacturer’s specifications
NucleoBond PC, mini kit for transfection-grade plasmid DNA (MACHEREY-NAGEL, REF 740571.100), store the components according to the manufacturer’s specifications
Blood & cell culture DNA mini kit (QIAGEN, catalog number: 13323), store the components according to the manufacturer’s specifications
RNeasy mini kit (QIAGEN, catalog number: 74004), store the components according to the manufacturer’s specifications
SuperScript® III first-strand synthesis system for RT-PCR (Invitrogen, catalog number: 18080-051), store the components according to the manufacturer’s specifications
iTaqTM Universal SYBR® green supermix (Bio-Rad, catalog number: 172-5121), store the components according to the manufacturer’s specifications
4–15% Criterion TGX precast midi protein gel (Bio-Rad, catalog number: 5671085), store at 4 °C
Trans-blot turbo midi 0.2 µm nitrocellulose membranes (Bio-Rad, catalog number: 1704159), store at 4 °C
Blotto, non-fat dry milk (Santa Cruz, catalog number: sc-2325), store at room temperature
Donkey anti-rabbit IgG (H+L), Alexa FluorTM Plus 800 (Thermo Fisher Scientific, catalog number: A32808)
Alexa Fluor® 680 AffiniPure donkey anti-sheep IgG (H+L) (Jackson ImmunoResearch, catalog number: 713-625-147)
1× PBS, store at room temperature
Growth media for HuTu-80 cells (see Recipes), store at 4 °C
Growth media for HEK 293T cells (see Recipes), store at 4 °C
Lysogeny broth (LB; Miller formulation, see Recipes), store at 4 °C
LB agar plates (see Recipes), store at 4 °C
10× Tris-buffered saline (TBS) solution (see Recipes)
Protein lysis buffer, store at 4 °C (see Recipes)
4× sample buffer, store at 4 °C (see Recipes)
Equipment
Thermal cycler (Bio-Rad, model: C1000TouchTM)
Horizontal gel electrophoresis system
Molecular Imager® GelDoc XR System (Bio-Rad)
3790 series genetic analyzer (Thermo Fisher Scientific)
Benchtop orbital incubator shaker (New Brunswick Scientific, model: Innova® 40/40R)
Eppendorf centrifuge 5810 equipped with the A-4-81 rotor (Eppendorf)
Cell culture CO2 incubator (Sanyo, model: MCO-19AIC)
Benchtop EVOS M5000 imaging system (AMF5000) equipped with a GFP light cube (AMEP4951)
Water bath (Precision – CIR 35, Fisher Scientific)
High sensitivity flow cytometer BD FACSAriaTM III (BD Biosciences)
CFX Opus 96 Real-Time PCR (Bio-Rad)
Heraeus FrescoTM 17 microcentrifuge equipped with 24 × 1.5/2.0 mL rotor with ClickSealTM biocontainment lid (Thermo Scientific)
Trans-Blot® TurboTM transfer system (Bio-Rad)
Odyssey CLx imaging system (LI-COR Biosciences)
Software
CHOPCHOP (version 3, https://chopchop.cbu.uib.no/)
TIDE: Tracking of Indels by Decomposition (http://shinyapps.datacurators.nl/tide/)
CFX Maestro Software for CFX Real-Time PCR Instruments (Bio-Rad)
Image StudioTM Lite Ver 5.2 (LI-COR Biosciences)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Grochowska, M. M., Bonifati, V. and Mandemakers, W. (2022). CRISPR/Cas9-mediated LRP10 Knockout in HuTu-80 and HEK 293T Cell Lines. Bio-protocol 12(19): e4521. DOI: 10.21769/BioProtoc.4521.
分类
细胞生物学 > 细胞工程 > CRISPR-cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link