- EN - English
- CN - 中文
Analysis of Lipid-linked Oligosaccharides Synthesized in vivo in Schizosaccharomyces pombe
裂殖酵母体内合成的脂联寡糖分析
(*contributed equally to this work) 发布: 2022年09月20日第12卷第18期 DOI: 10.21769/BioProtoc.4508 浏览次数: 2234
评审: Anonymous reviewer(s)
Abstract
Dolichol diphosphate-linked oligosaccharides (LLO) are the sugar donors in N-glycosylation, a fundamental protein post-translational modification of the eukaryotic secretory pathway. Defects in LLO biosynthesis produce human Congenital Disorders of Glycosylation Type I. The synthesis of LLOs and the transfer reactions to their protein acceptors is highly conserved among animal, plant, and fungi kingdoms, making the fission yeast Schizosaccharomyces pombe a suitable model to study these processes. Here, we present a protocol to determine the LLO patterns produced in vivo by S. pombe cells that may be easily adapted to other cell types. First, exponentially growing cultures are labeled with a pulse of [14C]-glucose. LLOs are then purified by successive extractions with organic solvents, and glycans are separated from the lipid moieties in mild acid hydrolysis and a new solvent extraction. The purified glycans are then run on paper chromatography. We use a deconvolution process to adjust the profile obtained to the minimal number of Gaussian functions needed to fit the data and determine the proportion of each species with respect to total glycan species present in the cell. The method we provide here might be used without any expensive or specialized equipment. The deconvolution process described here might also be useful to analyze species in non-completely resolved chromatograms.
Graphical abstract:
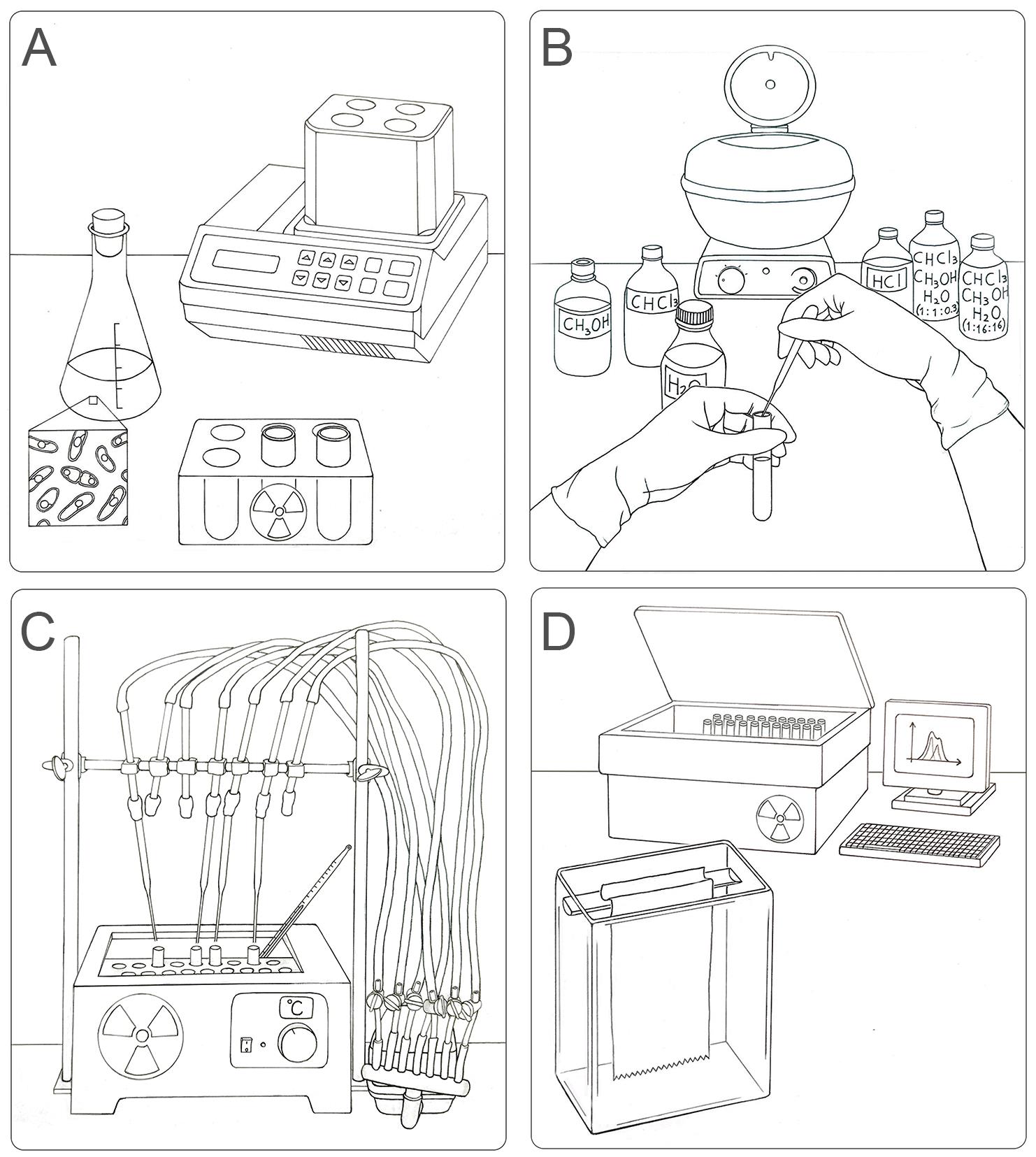
Workflow for the labeling, extraction, separation, and identification of LLO species in S. pombe. (A) Radioactive pulse of S. pombe cells with [14C]-glucose for 15 min at 28 °C. (B) Organic extraction of LLOs from labeled yeasts sequentially using methanol, chloroform, H2O, chloroform:methanol:H2O (1:1:0.3), 0.02 M HCl (to separate glycans from dolichol), and chloroform:methanol:H2O (1:16:16). (C) Preparation of the sample for chromatography on paper: drying by airflow and radioactivity check. (D) Loading of samples in chromatographic paper and descendent chromatography in a glass chamber. The obtained plots (CPM versus running distance) need to be analyzed to identify single glycan species.
Background
N-glycosylation is a frequent post-translational modification in the secretory pathway of eukaryotic cells involved in glycoprotein folding in the endoplasmic reticulum (ER). During N-glycosylation, a glycan composed of three glucoses (Glc), nine mannoses (Man), and two N-acetyl glucosamines (G3M9) is transferred in a single reaction step from a dolichol diphosphate oligosaccharide (LLO) donor to a consensus sequence present in proteins entering the ER (Cali et al., 2008). The reaction is catalyzed by the oligosaccharyltransferase (OST), which discriminates among completed and non-completed LLOs. The LLOs are synthesized stepwise in the ER membrane by different ALG gene products, first in the cytosolic side and then in the luminal side of the ER membrane (Figure 1) (Aebi, 2013; Stanley et al., 2015). Defects in LLOs synthesis result in their poor recognition by OST and thus less efficient transfer to proteins, leaving empty some normally occupied N-glycosylation sites. This hypoglycosylation of proteins may have profound effects in the cell and lead to several different Congenital Disorders of Glycosylation (CDG) type I (Ondruskova et al., 2021). These diseases are difficult to diagnose. The first confirmation usually arrives when hypoglycosylation is observed in serum transferrin, and a defect in LLO synthesis is assumed. To know which ALG genes are defective in each patient, either the biochemical analysis of LLO or a whole exome sequencing are required (Chang et al., 2018; Ng and Freeze, 2018; Wilson and Matthijs, 2021). The core machinery of LLO synthesis and N-glycosylation is highly conserved among eukaryotic species (Aebi, 2013), particularly between the fission yeast Schizosaccharomyces pombe and mammals (Fernandez et al., 1994). S. pombe is easily handled, facilities required for its growth and maintenance are inexpensive, and many biochemical and genetic tools are already developed for this organism. For these reasons, S. pombe has become a suitable model organism for research, and several molecular tools developed and optimized for use in this model organism have been used in mammalian cells (Hagan, 2016).
We present here a detailed protocol to specifically identify the LLO species synthesized in S. pombe in vivo. The key step of the protocol is to stop protein synthesis with inhibitors preventing glycan transfer to proteins while radioactive labeled LLOs are being synthesized. Cultures are compared under similar nutritional growth conditions, in order to avoid differences in LLO structures due to environmental conditions or metabolic constraints (Gershman and Robbins, 1981; Chapman and Calhoun, 1988). We then extract LLOs from the labeled cells and analyze them either by HPLC or by paper chromatography. To better identify the peaks obtained, we developed a deconvolution process using a Gaussian function available in a Microsoft Excel Solver datasheet provided here. This method allows a better visualization of the peaks, making them easily comparable with the standards run in parallel, and provides an accurate quantification of each peak. With the protocol described here, we were able to distinguish the LLOs produced in 16 S. pombe strains that synthesize combinatorial possible structures of LLOs (GXMY, where X is the number of Glc residues in the glycan, ranging from three to zero, and Y is the number of Man units, ranging from nine to five) (Gallo et al., 2022). This low-cost method has the advantages of being readily available for almost all laboratories, providing high accuracy, and being easily adapted to any cell type, simplifying the determination of the LLO biosynthetic step blocked in a particular cell mutant or synthesized by the organism being studied. Moreover, the method we provide to obtain the deconvolution of the LLO profiles may be used for other chromatographic resolutions.
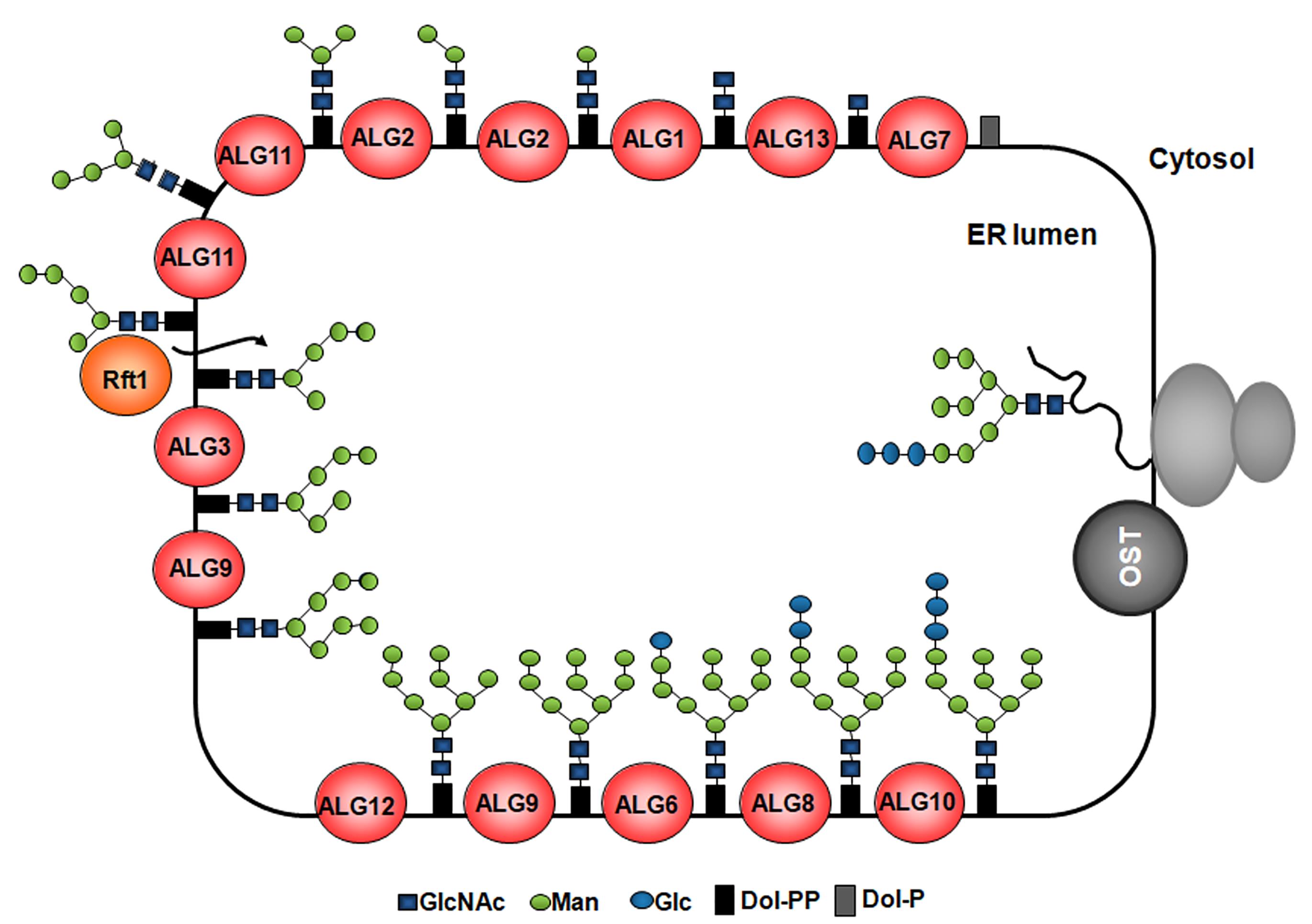
Figure 1. Synthesis of LLOs and glycan transfer to nascent proteins in the ER. The G3M9 LLO is built in the ER membrane step-by-step by monosaccharide addition mediated by specific glycosyltransferases encoded by alg (asparagine-linked glycosylation) genes. Two N-acetylglucosamines and five Man are transferred to the dolichol from nucleotide-sugars on the cytoplasmic side of the ER membrane. Then, the LLO intermediate bearing five Man residues is flipped to the ER lumen, where four additional Man residues are added by the ALG3-, ALG9-, and ALG12-encoded ER luminal dolichol-dependent mannosyltransferases. Finally, three Glc residues are incorporated by the ALG6-, ALG8-, and ALG10-encoded dolichol-dependent glucosyltransferases. Once completed, the LLO is transferred by OST to nascent polypeptides entering the ER through the translocon.
Materials and Reagents
Materials
Conical tubes, 50 mL (Biologix, catalog number: 10-9502)
Polypropylene centrifuge bottle with Screw-On Cap, 250 mL (Beckman Coulter, catalog number: 356011)
Polypropylene centrifuge bottle with Screw-On Cap, 50 mL (Beckman Coulter, catalog number: 357003)
Metal spatula
Glass beads 0.5 mm diameter (Millipore Sigma, Sigma, catalog number: G8772)
Glass tubes (Kimax, catalog number: 45048)
Glass eyedropper
Chromatography paper sheets (Sartorius Stedim FN3 580 × 600 mm)
Scintillation 20 mL glass counting vials (Wheaton, catalog number: 986532)
Reagents
Yeast extract (BactoTM, catalog number: 212750), store at room temperature (RT)
Adenine (Millipore Sigma, Sigma, catalog number: A8626), store at RT
Yeast nitrogen base (YNB) (Thermo Fisher Scientific, BD Difco, catalog number: DF0392-15-9), store at RT
Puromycin 10 mg/mL (InvivoGen, catalog number: ant-pr-1), store at 4 °C
Cycloheximide 2 mg/mL (Millipore Sigma, catalog number: C-6255), store at -20 °C
D-[14C(U)]-Glc (Perkin Elmer, catalog number: NEC042V20UC), store at 4 °C
Glucose 1 M (Dextrose 1-hydrate, Biopack, catalog number: 9812.08), store at RT
Methanol 99.9% (Millipore Sigma, Merck, catalog number: 106009), store at RT
Chloroform (Merck, catalog number: 107024), store at RT
1-propanol 98% (Millipore Sigma, Merk, catalog number: 100997), store at RT
Nitromethane (Millipore Sigma, catalog number: 270423), store at RT
Hydrochloric acid fuming 37% for analysis (Merck, catalog number: 1003171000)
PPO-POPOP scintillation cocktail (concentrated scintillator PPO-POPOP, RPI, catalog number: 111045)
Milli Q water
Yeast extract medium supplemented with adenine (YEA) (see Recipes)
Yeast nitrogen base (YNB) without glucose (see Recipes)
Radioactive solution to label a 500 μL sample (see Recipes)
Chloroform:methanol:water (1:1:0.3) (see Recipes)
Chloroform:methanol:water (1:16:16) (see Recipes)
0.02 N HCl (see Recipes)
Equipment
1 L glass flask
Orbital shaker (for flasks) set at 28 °C (Forma Scientific, model: 4581)
OD600 reader (Amersham Biosciences, model: Ultraspec 10)
Centrifuge (Beckman Avanti) with rotors JA-14 and JA-20
Thermomixer set at 28 °C (Thermo-shaker, mrc)
Water bath (Vicking SRL)
Airflow (Aquarium air pump RS-Electric, model: RS-15000)
Analog benchtop centrifuge (Rolco)
Vortex (Scientific Industries, model: Vortex-genie 2)
Freezer -20 °C (KENT)
Glass chromatography chamber (Homemade)
Scintillation counter (Pharmacia, RackBeta 1214/1219)
NOTE: The equipment described here may easily be replaced for common lab equipment of other brands.
Software
Microsoft Excel (version 2016) (Microsoft.com)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Valko, A., Gallo, G. L., Weisz, A. D., Parodi, A. J. and D'Alessio, C. (2022). Analysis of Lipid-linked Oligosaccharides Synthesized in vivo in Schizosaccharomyces pombe. Bio-protocol 12(18): e4508. DOI: 10.21769/BioProtoc.4508.
分类
微生物学 > 微生物生物化学 > 糖类
微生物学 > 微生物生物化学 > 脂质
生物化学 > 糖类 > 寡糖 > 标记
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










![N-连接寡糖的[2-<sup>3</sup>H] 甘露糖标记和分析](https://en-cdn.bio-protocol.org/imageup/arcimg/20170718074630401.jpg?t=1771668024)

